[English] 日本語
 Yorodumi
Yorodumi- EMDB-8268: HIV NFL trimer cross-linked with Glutaraldehyde and in complex wi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8268 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
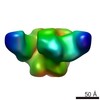
| Title | HIV NFL trimer cross-linked with Glutaraldehyde and in complex with bnAb VRC01 | |||||||||
 Map data Map data | HIV NFL trimer cross-linked with Glutaraldehyde and in complex with bnAb VRC01 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 20.0 Å | |||||||||
 Authors Authors | Ward AB / de Val N | |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2016 Journal: PLoS Pathog / Year: 2016Title: Thermostability of Well-Ordered HIV Spikes Correlates with the Elicitation of Autologous Tier 2 Neutralizing Antibodies. Authors: Yu Feng / Karen Tran / Shridhar Bale / Shailendra Kumar / Javier Guenaga / Richard Wilson / Natalia de Val / Heather Arendt / Joanne DeStefano / Andrew B Ward / Richard T Wyatt /  Abstract: In the context of HIV vaccine design and development, HIV-1 spike mimetics displaying a range of stabilities were evaluated to determine whether more stable, well-ordered trimers would more ...In the context of HIV vaccine design and development, HIV-1 spike mimetics displaying a range of stabilities were evaluated to determine whether more stable, well-ordered trimers would more efficiently elicit neutralizing antibodies. To begin, in vitro analysis of trimers derived from the cysteine-stabilized SOSIP platform or the uncleaved, covalently linked NFL platform were evaluated. These native-like trimers, derived from HIV subtypes A, B, and C, displayed a range of thermostabilities, and were "stress-tested" at varying temperatures as a prelude to in vivo immunogenicity. Analysis was performed both in the absence and in the presence of two different adjuvants. Since partial trimer degradation was detected at 37°C before or after formulation with adjuvant, we sought to remedy such an undesirable outcome. Cross-linking (fixing) of the well-ordered trimers with glutaraldehyde increased overall thermostability, maintenance of well-ordered trimer integrity without or with adjuvant, and increased resistance to solid phase-associated trimer unfolding. Immunization of unfixed and fixed well-ordered trimers into animals revealed that the elicited tier 2 autologous neutralizing activity correlated with overall trimer thermostability, or melting temperature (Tm). Glutaraldehyde fixation also led to higher tier 2 autologous neutralization titers. These results link retention of trimer quaternary packing with elicitation of tier 2 autologous neutralizing activity, providing important insights for HIV-1 vaccine design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8268.map.gz emd_8268.map.gz | 1.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8268-v30.xml emd-8268-v30.xml emd-8268.xml emd-8268.xml | 11.4 KB 11.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8268.png emd_8268.png | 16.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8268 http://ftp.pdbj.org/pub/emdb/structures/EMD-8268 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8268 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8268 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8268.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8268.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HIV NFL trimer cross-linked with Glutaraldehyde and in complex with bnAb VRC01 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : BG505_NFL_trimer cross-linked with glutaraldehyde and in complex ...
| Entire | Name: BG505_NFL_trimer cross-linked with glutaraldehyde and in complex with VRC01 bnAb |
|---|---|
| Components |
|
-Supramolecule #1: BG505_NFL_trimer cross-linked with glutaraldehyde and in complex ...
| Supramolecule | Name: BG505_NFL_trimer cross-linked with glutaraldehyde and in complex with VRC01 bnAb type: complex / ID: 1 / Parent: 0 Details: BG505_NFL_trimer cross-linked with glutaraldehyde and in complex with 3 broadly neutralizing antibodies VRC01 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK 293F / Recombinant plasmid: pPPI4 Homo sapiens (human) / Recombinant cell: HEK 293F / Recombinant plasmid: pPPI4 |
| Molecular weight | Theoretical: 570 KDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||
| Staining | Type: NEGATIVE / Material: Uranyl Formate Details: Sample was stained using two applications of 3 uL 2% UF. | |||||||||
| Grid | Model: Electron Microscopy Sciences / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Details: The grids were glow-discharged at 20 mA. | |||||||||
| Details | The trimer was incubated at RT with a 10 M excess of the broadly neutralizing antibody VRC01. The complex was purified by SEC. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number grids imaged: 1 / Number real images: 60 / Average electron dose: 36.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus min: 1.0 µm / Nominal magnification: 46000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















