-Search query
-Search result
Showing 1 - 50 of 1,079 items for (author: cui & n)

EMDB-37441: 
FCP tetramer in Chaetoceros gracilis

EMDB-37442: 
FCP pentamer in Chaetoceros gracilis

PDB-8wck: 
FCP tetramer in Chaetoceros gracilis

PDB-8wcl: 
FCP pentamer in Chaetoceros gracilis

EMDB-39645: 
The structure of HKU1-B S protein with bsAb1

EMDB-39646: 
the complex structure of the H4B6 Fab with the RBD of Omicron BA.5 S protein

PDB-8yww: 
The structure of HKU1-B S protein with bsAb1

PDB-8ywx: 
the complex structure of the H4B6 Fab with the RBD of Omicron BA.5 S protein

EMDB-45364: 
HIV-1 intasome core bound with DTG

EMDB-38617: 
SARS-CoV-2 RBD + IMCAS-123 + IMCAS-72 Fab

EMDB-38618: 
SARS-CoV-2 RBD + IMCAS-364 + hACE2

EMDB-38619: 
SARS-CoV-2 RBD + IMCAS-364 (Local Refinement)

EMDB-38620: 
SARS-CoV-2 Omicron BA.4 RBD + IMCAS-316 + ACE2

EMDB-38621: 
SARS-CoV-2 spike + IMCAS-123

EMDB-38823: 
Cryo-EM structure of the 123-316 scDb/PT-RBD complex

PDB-8xse: 
SARS-CoV-2 RBD + IMCAS-123 + IMCAS-72 Fab

PDB-8xsf: 
SARS-CoV-2 RBD + IMCAS-364 + hACE2

PDB-8xsi: 
SARS-CoV-2 RBD + IMCAS-364 (Local Refinement)

PDB-8xsj: 
SARS-CoV-2 Omicron BA.4 RBD + IMCAS-316 + ACE2

PDB-8xsl: 
SARS-CoV-2 spike + IMCAS-123

PDB-8y0y: 
Cryo-EM structure of the 123-316 scDb/PT-RBD complex

EMDB-16384: 
Tetrameric 5-HT3A receptor in Salipro (apo, asymmetric)

EMDB-16385: 
Tetrameric 5-HT3aR in Salipro (apo state, symmetric)

EMDB-16386: 
Tetrameric 5-HT3aR in Salipro (holo state, symmetric)

EMDB-16387: 
Tetrameric 5-HT3A receptor in Salipro (holo, asymmetric)

EMDB-36946: 
Dimer structure of procaryotic Ago

EMDB-36947: 
Structure of procaryotic Ago

PDB-8k87: 
Dimer structure of procaryotic Ago

PDB-8k88: 
Structure of procaryotic Ago

EMDB-36466: 
FCP trimer in diatom Thalassiosira pseudonana

PDB-8jp3: 
FCP trimer in diatom Thalassiosira pseudonana

EMDB-38156: 
Structure of enterovirus protease in complex host factor

PDB-8x8q: 
Structure of enterovirus protease in complex host factor

EMDB-36721: 
Structure of TbAQP2 in complex with anti-trypanosomatid drug melarsoprol

EMDB-36722: 
Structure of the TbAQP2 in the apo conformation

EMDB-36723: 
Structure of TbAQP2 in complex with anti-trypanosomatid drug pentamidine

PDB-8jy6: 
Structure of TbAQP2 in complex with anti-trypanosomatid drug melarsoprol

PDB-8jy7: 
Structure of the TbAQP2 in the apo conformation

PDB-8jy8: 
Structure of TbAQP2 in complex with anti-trypanosomatid drug pentamidine

EMDB-38394: 
The Cryo-EM structure of MPXV E5 apo conformation

EMDB-38395: 
The Cryo-EM structure of MPXV E5 in complex with DNA

EMDB-38396: 
The Cryo-EM structure of MPXV E5 C-terminal in complex with DNA

PDB-8xj6: 
The Cryo-EM structure of MPXV E5 apo conformation

PDB-8xj7: 
The Cryo-EM structure of MPXV E5 in complex with DNA

PDB-8xj8: 
The Cryo-EM structure of MPXV E5 C-terminal in complex with DNA
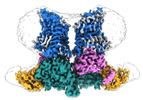
EMDB-42014: 
Structural Basis of Human NOX5 Activation
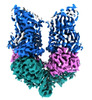
EMDB-42015: 
Structural Basis of Human NOX5 Activation
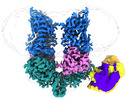
EMDB-42016: 
Structural Basis of Human NOX5 Activation

EMDB-42345: 
Structural Basis of Human NOX5 Activation

EMDB-42348: 
Structural Basis of Human NOX5 Activation
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
