+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8711 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Phage Qbeta in complex with MurA | |||||||||||||||
 Map data Map data | Enterobacteria phage Qbeta virion with MurA | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Qbeta /  ssRNA / ssRNA /  phage / phage /  virus / VIRUS-TRANSFERASE complex virus / VIRUS-TRANSFERASE complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsuppression by virus of host cell wall biogenesis / suppression by virus of host peptidoglycan biosynthetic process / viral release via suppression of host peptidoglycan biosynthetic process / virion attachment to host cell pilus /  UDP-N-acetylglucosamine 1-carboxyvinyltransferase / UDP-N-acetylglucosamine 1-carboxyvinyltransferase /  UDP-N-acetylglucosamine 1-carboxyvinyltransferase activity / UDP-N-acetylgalactosamine biosynthetic process / adhesion receptor-mediated virion attachment to host cell / peptidoglycan biosynthetic process / viral release from host cell by cytolysis ...suppression by virus of host cell wall biogenesis / suppression by virus of host peptidoglycan biosynthetic process / viral release via suppression of host peptidoglycan biosynthetic process / virion attachment to host cell pilus / UDP-N-acetylglucosamine 1-carboxyvinyltransferase activity / UDP-N-acetylgalactosamine biosynthetic process / adhesion receptor-mediated virion attachment to host cell / peptidoglycan biosynthetic process / viral release from host cell by cytolysis ...suppression by virus of host cell wall biogenesis / suppression by virus of host peptidoglycan biosynthetic process / viral release via suppression of host peptidoglycan biosynthetic process / virion attachment to host cell pilus /  UDP-N-acetylglucosamine 1-carboxyvinyltransferase / UDP-N-acetylglucosamine 1-carboxyvinyltransferase /  UDP-N-acetylglucosamine 1-carboxyvinyltransferase activity / UDP-N-acetylgalactosamine biosynthetic process / adhesion receptor-mediated virion attachment to host cell / peptidoglycan biosynthetic process / viral release from host cell by cytolysis / UDP-N-acetylglucosamine 1-carboxyvinyltransferase activity / UDP-N-acetylgalactosamine biosynthetic process / adhesion receptor-mediated virion attachment to host cell / peptidoglycan biosynthetic process / viral release from host cell by cytolysis /  virion component / cell wall organization / regulation of cell shape / virion component / cell wall organization / regulation of cell shape /  cell cycle / symbiont entry into host cell / cell cycle / symbiont entry into host cell /  cell division / cell division /  cytoplasm cytoplasmSimilarity search - Function | |||||||||||||||
| Biological species |   Escherichia coli O139:H28 (strain E24377A / ETEC) (bacteria) / Escherichia coli O139:H28 (strain E24377A / ETEC) (bacteria) /   Escherichia phage Qbeta (virus) / Escherichia phage Qbeta (virus) /   Enterobacteria phage Qbeta (virus) Enterobacteria phage Qbeta (virus) | |||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 5.7 Å cryo EM / Resolution: 5.7 Å | |||||||||||||||
 Authors Authors | Cui Z / Zhang J | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2017 Journal: Proc Natl Acad Sci U S A / Year: 2017Title: Structures of Qβ virions, virus-like particles, and the Qβ-MurA complex reveal internal coat proteins and the mechanism of host lysis. Authors: Zhicheng Cui / Karl V Gorzelnik / Jeng-Yih Chang / Carrie Langlais / Joanita Jakana / Ry Young / Junjie Zhang /  Abstract: In single-stranded RNA bacteriophages (ssRNA phages) a single copy of the maturation protein binds the genomic RNA (gRNA) and is required for attachment of the phage to the host pilus. For the ...In single-stranded RNA bacteriophages (ssRNA phages) a single copy of the maturation protein binds the genomic RNA (gRNA) and is required for attachment of the phage to the host pilus. For the canonical Qβ the maturation protein, A, has an additional role as the lysis protein, by its ability to bind and inhibit MurA, which is involved in peptidoglycan biosynthesis. Here, we determined structures of Qβ virions, virus-like particles, and the Qβ-MurA complex using single-particle cryoelectron microscopy, at 4.7-Å, 3.3-Å, and 6.1-Å resolutions, respectively. We identified the outer surface of the β-region in A as the MurA-binding interface. Moreover, the pattern of MurA mutations that block Qβ lysis and the conformational changes of MurA that facilitate A binding were found to be due to the intimate fit between A and the region encompassing the closed catalytic cleft of substrate-liganded MurA. Additionally, by comparing the Qβ virion with Qβ virus-like particles that lack a maturation protein, we observed a structural rearrangement in the capsid coat proteins that is required to package the viral gRNA in its dominant conformation. Unexpectedly, we found a coat protein dimer sequestered in the interior of the virion. This coat protein dimer binds to the gRNA and interacts with the buried α-region of A, suggesting that it is sequestered during the early stage of capsid formation to promote the gRNA condensation required for genome packaging. These internalized coat proteins are the most asymmetrically arranged major capsid proteins yet observed in virus structures. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8711.map.gz emd_8711.map.gz | 15.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8711-v30.xml emd-8711-v30.xml emd-8711.xml emd-8711.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8711.png emd_8711.png | 221.4 KB | ||
| Filedesc metadata |  emd-8711.cif.gz emd-8711.cif.gz | 5.9 KB | ||
| Others |  emd_8711_additional.map.gz emd_8711_additional.map.gz | 41.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8711 http://ftp.pdbj.org/pub/emdb/structures/EMD-8711 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8711 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8711 | HTTPS FTP |
-Related structure data
| Related structure data |  5vm7MC  8707C  8708C  8709C  8710C  5vlyC  5vlzC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8711.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8711.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Enterobacteria phage Qbeta virion with MurA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.25 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Enterobacteria phage Qbeta virion with MurA, additional map
| File | emd_8711_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Enterobacteria phage Qbeta virion with MurA, additional map | ||||||||||||
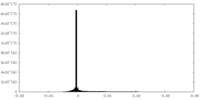
| Projections & Slices |
| ||||||||||||
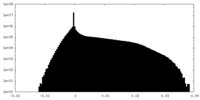
| Density Histograms |
- Sample components
Sample components
-Entire : Enterobacteria phage Qbeta virion with MurA
| Entire | Name: Enterobacteria phage Qbeta virion with MurA |
|---|---|
| Components |
|
-Supramolecule #1: Enterobacteria phage Qbeta virion with MurA
| Supramolecule | Name: Enterobacteria phage Qbeta virion with MurA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #3: UDP-N-acetylglucosamine 1-carboxyvinyltransferase
| Supramolecule | Name: UDP-N-acetylglucosamine 1-carboxyvinyltransferase / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:   Escherichia coli O139:H28 (strain E24377A / ETEC) (bacteria) Escherichia coli O139:H28 (strain E24377A / ETEC) (bacteria) |
-Supramolecule #2: Enterobacteria phage Qbeta
| Supramolecule | Name: Enterobacteria phage Qbeta / type: virus / ID: 2 / Parent: 1 / Macromolecule list: #1 / NCBI-ID: 39803 / Sci species name: Enterobacteria phage Qbeta / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Maturation protein A2
| Macromolecule | Name: Maturation protein A2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia phage Qbeta (virus) Escherichia phage Qbeta (virus) |
| Molecular weight | Theoretical: 48.613078 KDa |
| Sequence | String: MPKLPRGLRF GADNEILNDF QELWFPDLFI ESSDTHPWYT LKGRVLNAHL DDRLPNVGGR QVRRTPHRVT VPIASSGLRP VTTVQYDPA ALSFLLNARV DWDFGNGDSA NLVINDFLFR TFAPKEFDFS NSLVPRYTQA FSAFNAKYGT MIGEGLETIK Y LGLLLRRL ...String: MPKLPRGLRF GADNEILNDF QELWFPDLFI ESSDTHPWYT LKGRVLNAHL DDRLPNVGGR QVRRTPHRVT VPIASSGLRP VTTVQYDPA ALSFLLNARV DWDFGNGDSA NLVINDFLFR TFAPKEFDFS NSLVPRYTQA FSAFNAKYGT MIGEGLETIK Y LGLLLRRL REGYRAVKRG DLRALRRVIQ SYHNGKWKPA TAGNLWLEFR YGLMPLFYDI RDVMLDWQNR HDKIQRLLRF SV GHGEDYV VEFDNLYPAV AYFKLKGEIT LERRHRHGIS YANREGYAVF DNGSLRPVSD WKELATAFIN PHEVAWELTP YSF VVDWFL NVGDILAQQG QLYHNIDIVD GFDRRDIRLK SFTIKGERNG RPVNVSASLS AVDLFYSRLH TSNLPFATLD LDTT FSSFK HVLDSIFLLT QRVKR UniProtKB: Maturation protein A2 |
-Macromolecule #2: UDP-N-acetylglucosamine 1-carboxyvinyltransferase
| Macromolecule | Name: UDP-N-acetylglucosamine 1-carboxyvinyltransferase / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number:  UDP-N-acetylglucosamine 1-carboxyvinyltransferase UDP-N-acetylglucosamine 1-carboxyvinyltransferase |
|---|---|
| Source (natural) | Organism:   Escherichia coli O139:H28 (strain E24377A / ETEC) (bacteria) Escherichia coli O139:H28 (strain E24377A / ETEC) (bacteria)Strain: E24377A / ETEC |
| Molecular weight | Theoretical: 44.871543 KDa |
| Sequence | String: MDKFRVQGPT KLQGEVTISG AKNAALPILF AALLAEEPVE IQNVPKLKDV DTSMKLLSQL GAKVERNGSV HIDARDVNVF CAPYDLVKT MRASIWALGP LVARFGQGQV SLPGGCTIGA RPVDLHISGL EQLGATIKLE EGYVKASVDG RLKGAHIVMD K VSVGATVT ...String: MDKFRVQGPT KLQGEVTISG AKNAALPILF AALLAEEPVE IQNVPKLKDV DTSMKLLSQL GAKVERNGSV HIDARDVNVF CAPYDLVKT MRASIWALGP LVARFGQGQV SLPGGCTIGA RPVDLHISGL EQLGATIKLE EGYVKASVDG RLKGAHIVMD K VSVGATVT IMCAATLAEG TTIIENAARE PEIVDTANFL ITLGAKISGQ GTDRIVIEGV ERLGGGVYRV LPDRIETGTF LV AAAISRG KIICRNAQPD TLDAVLAKLR DAGADIEVGE DWISLDMHGK RPKAVNVRTA PHPAFPTDMQ AQFTLLNLVA EGT GFITET VFENRFMHVP ELSRMGAHAE IESNTVICHG VEKLSGAQVM ATDLRASASL VLAGCIAEGT TVVDRIYHID RGYE RIEDK LRALGANIER VKGE UniProtKB:  UDP-N-acetylglucosamine 1-carboxyvinyltransferase UDP-N-acetylglucosamine 1-carboxyvinyltransferase |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK III Details: Blotted for 6s, Plunged into liquid ethane (FEI VITROBOT MARK III). |
- Electron microscopy #1
Electron microscopy #1
| Microscope | JEOL 3200FSC |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal magnification: 30000 Bright-field microscopy / Nominal magnification: 30000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Microscopy ID | 1 |
| Image recording | Image recording ID: 1 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 0.2 sec. / Average electron dose: 1.0 e/Å2 |
- Electron microscopy #1~
Electron microscopy #1~
| Microscope | FEI TECNAI F20 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal magnification: 29000 Bright-field microscopy / Nominal magnification: 29000 |
| Microscopy ID | 1 |
| Image recording | Image recording ID: 2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 0.2 sec. / Average electron dose: 1.1 e/Å2 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 5.7 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 1.4) / Number images used: 25597 |
| Image recording ID | 1 |
 Movie
Movie Controller
Controller















 Z
Z Y
Y X
X