[English] 日本語
 Yorodumi
Yorodumi- EMDB-3729: RNA polymerase I pre-initiation complex (Pol-Rrn3 focused refinement) -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3729 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
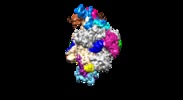
| Title | RNA polymerase I pre-initiation complex (Pol-Rrn3 focused refinement) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.85 Å cryo EM / Resolution: 3.85 Å | |||||||||
 Authors Authors | Sadian Y / Tafur L / Kosinski J / Jakobi AJ / Wetzel R / Buczak K / Hagen WJH / Beck M / Sachse C / Muller CW | |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2017 Journal: EMBO J / Year: 2017Title: Structural insights into transcription initiation by yeast RNA polymerase I. Authors: Yashar Sadian / Lucas Tafur / Jan Kosinski / Arjen J Jakobi / Rene Wetzel / Katarzyna Buczak / Wim Jh Hagen / Martin Beck / Carsten Sachse / Christoph W Müller /  Abstract: In eukaryotic cells, RNA polymerase I (Pol I) synthesizes precursor ribosomal RNA (pre-rRNA) that is subsequently processed into mature rRNA. To initiate transcription, Pol I requires the assembly of ...In eukaryotic cells, RNA polymerase I (Pol I) synthesizes precursor ribosomal RNA (pre-rRNA) that is subsequently processed into mature rRNA. To initiate transcription, Pol I requires the assembly of a multi-subunit pre-initiation complex (PIC) at the ribosomal RNA promoter. In yeast, the minimal PIC includes Pol I, the transcription factor Rrn3, and Core Factor (CF) composed of subunits Rrn6, Rrn7, and Rrn11. Here, we present the cryo-EM structure of the 18-subunit yeast Pol I PIC bound to a transcription scaffold. The cryo-EM map reveals an unexpected arrangement of the DNA and CF subunits relative to Pol I. The upstream DNA is positioned differently than in any previous structures of the Pol II PIC. Furthermore, the TFIIB-related subunit Rrn7 also occupies a different location compared to the Pol II PIC although it uses similar interfaces as TFIIB to contact DNA. Our results show that although general features of eukaryotic transcription initiation are conserved, Pol I and Pol II use them differently in their respective transcription initiation complexes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3729.map.gz emd_3729.map.gz | 2.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3729-v30.xml emd-3729-v30.xml emd-3729.xml emd-3729.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3729.png emd_3729.png | 111 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3729 http://ftp.pdbj.org/pub/emdb/structures/EMD-3729 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3729 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3729 | HTTPS FTP |
-Related structure data
| Related structure data |  3727C  3728C  5oa1FC F: fitted*YM C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3729.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3729.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : RNA polymerase I pre-initiation complex with Core Factor, Rrn3 an...
+Supramolecule #1: RNA polymerase I pre-initiation complex with Core Factor, Rrn3 an...
+Macromolecule #1: RPA1_YEAST DNA-directed RNA polymerase I subunit RPA190
+Macromolecule #2: RPA2_YEAST DNA-directed RNA polymerase I subunit RPA135
+Macromolecule #3: RPA49_YEAST DNA-directed RNA polymerase I subunit RPA49
+Macromolecule #4: RPA43_YEAST DNA-directed RNA polymerase I subunit RPA43
+Macromolecule #5: RPAC1_YEAST DNA-directed RNA polymerases I and III subunit RPAC1
+Macromolecule #6: RPA34_YEAST DNA-directed RNA polymerase I subunit RPA34
+Macromolecule #7: RPAB1_YEAST DNA-directed RNA polymerases I, II, and III subunit RPABC1
+Macromolecule #8: RPAB2_YEAST DNA-directed RNA polymerases I, II, and III subunit RPABC2
+Macromolecule #9: RPAC2_YEAST DNA-directed RNA polymerases I and III subunit RPAC2
+Macromolecule #10: RPAB3_YEAST DNA-directed RNA polymerases I, II, and III subunit RPABC3
+Macromolecule #11: RPA14_YEAST DNA-directed RNA polymerase I subunit RPA14
+Macromolecule #12: RPA12_YEAST DNA-directed RNA polymerase I subunit RPA12
+Macromolecule #13: RPAB4_YEAST DNA-directed RNA polymerases I, II, and III subunit RPABC4
+Macromolecule #14: RPAB5_YEAST DNA-directed RNA polymerases I, II, and III subunit RPABC5
+Macromolecule #15: RRN3_YEAST RNA polymerase I-specific transcription initiation fac...
+Macromolecule #16: non-template DNA strand
+Macromolecule #17: template DNA strand
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 105000 Bright-field microscopy / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 105000 |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 4235 / Average exposure time: 20.0 sec. / Average electron dose: 2.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Software - Name: CTFFIND (ver. 4) |
|---|---|
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.85 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 1.4) / Number images used: 38589 |
 Movie
Movie Controller
Controller