+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | TSWV L protein in complex with ribavirin 5-triphosphate | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TSWV / L protein / ribavirin 5-triphosphate / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA-directed RNA polymerase / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription Similarity search - Function | |||||||||
| Biological species |  Orthotospovirus tomatomaculae Orthotospovirus tomatomaculae | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Cao L / Wang X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2025 Journal: Nat Plants / Year: 2025Title: Structural basis for the activation of plant bunyavirus replication machinery and its dual-targeted inhibition by ribavirin. Authors: Jia Li / Lei Cao / Yaqian Zhao / Jinghan Shen / Lei Wang / Mingfeng Feng / Min Zhu / Yonghao Ye / Richard Kormelink / Xiaorong Tao / Xiangxi Wang /   Abstract: Despite the discovery of plant viruses as a new class of pathogens over a century ago, the structure of plant virus replication machinery and antiviral pesticide remains lacking. Here we report five ...Despite the discovery of plant viruses as a new class of pathogens over a century ago, the structure of plant virus replication machinery and antiviral pesticide remains lacking. Here we report five cryogenic electron microscopy structures of a ~330-kDa RNA-dependent RNA polymerase (RdRp) from a devastating plant bunyavirus, tomato spotted wilt orthotospovirus (TSWV), including the apo, viral-RNA-bound, base analogue ribavirin-bound and ribavirin-triphosphate-bound states. They reveal that a flexible loop of RdRp's motif F functions as 'sensor' to perceive viral RNA and further acts as an 'adaptor' to promote the formation of a complete catalytic centre. A ten-base RNA 'hook' structure is sufficient to trigger major conformational changes and activate RdRp. Chemical screening showed that ribavirin is effective against TSWV, and structural data revealed that ribavirin disrupts both hook-binding and catalytic core formation, locking polymerase in its inactive state. This work provides structural insights into the mechanisms of plant bunyavirus RdRp activation and its dual-targeted site inhibition, facilitating the development of pesticides against plant viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61241.map.gz emd_61241.map.gz | 51.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61241-v30.xml emd-61241-v30.xml emd-61241.xml emd-61241.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_61241.png emd_61241.png | 74.2 KB | ||
| Filedesc metadata |  emd-61241.cif.gz emd-61241.cif.gz | 6.4 KB | ||
| Others |  emd_61241_half_map_1.map.gz emd_61241_half_map_1.map.gz emd_61241_half_map_2.map.gz emd_61241_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61241 http://ftp.pdbj.org/pub/emdb/structures/EMD-61241 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61241 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61241 | HTTPS FTP |
-Validation report
| Summary document |  emd_61241_validation.pdf.gz emd_61241_validation.pdf.gz | 884.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_61241_full_validation.pdf.gz emd_61241_full_validation.pdf.gz | 884.2 KB | Display | |
| Data in XML |  emd_61241_validation.xml.gz emd_61241_validation.xml.gz | 13.3 KB | Display | |
| Data in CIF |  emd_61241_validation.cif.gz emd_61241_validation.cif.gz | 15.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61241 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61241 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61241 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61241 | HTTPS FTP |
-Related structure data
| Related structure data |  9j8vMC  8ki6C  8ki7C  8ki8C  8ki9C  8kiaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_61241.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61241.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_61241_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_61241_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TSWV L protein
| Entire | Name: TSWV L protein |
|---|---|
| Components |
|
-Supramolecule #1: TSWV L protein
| Supramolecule | Name: TSWV L protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Orthotospovirus tomatomaculae Orthotospovirus tomatomaculae |
-Macromolecule #1: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Orthotospovirus tomatomaculae Orthotospovirus tomatomaculae |
| Molecular weight | Theoretical: 203.180734 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: FFSHWTSKYK ERNPTEIAYS EDIERIIDSL VTDEITKEEI IHFLFGNFCF HIETMNDQHI ADKFKGYQSS CINLKIEPKV DLADLKDHL IQKQQIWESL YGKHLEKIML RIREKKKKEK EIPDITTAFN QNAAEYEEKY PNCFTNDLSE TKTNFSMTWS P SFEKIELS ...String: FFSHWTSKYK ERNPTEIAYS EDIERIIDSL VTDEITKEEI IHFLFGNFCF HIETMNDQHI ADKFKGYQSS CINLKIEPKV DLADLKDHL IQKQQIWESL YGKHLEKIML RIREKKKKEK EIPDITTAFN QNAAEYEEKY PNCFTNDLSE TKTNFSMTWS P SFEKIELS SEVDYNNAII NKFRESFKSS SRVIYNSPYS SINNQTNKAR DITNLVRLCL TELSCDTTKM EKQELEDEID IN TGSIKVE RTKKSKEWNK QGSCLTRNKN EFCMKETGRE NKTIYFKGLA VMNIGMSSKK RILKKEEIKE RISKGLEYDT SER QADPND DYSSIDMSSL THMKKLIRHD NEDSLSWCER IKDSLFVLHN GDIREEGKIT SVYNNYAKNP ECLYIQDSVL KTEL ETCKK INKLCNDLAI YHYSEDMMQF SKGLMVADRY MTKESFKILT TANTSMMLLA FKGDGMNTGG SGVPYIALHI VDEDM SDQF NICYTKEIYS YFRNGSNYIY IMRPQRLNQV RLLSLFKTPS KVPVCFAQFS KKANEMEKWL KNKDIEKVNV FSMTMT VKQ ILINIVFSSV MIGTVTKLSR MGIFDFMRYA GFLPLSDYSN IKEYIRDKFD PDITNVADIY FVNGIKKLLF RMEDLNL ST NAKPVVVDHE NDIIGGITDL NIKCPITGST LLTLEDLYNN VYLAIYMMPK SLHNHVHNLT SLLNVPAEWE LKFRKELG F NIFEDIYPKK AMFDDKDLFS INGALNVKAL SDYYLGNIEN VGLMRSEIEN KEDFLSPCYK ISTLKSSKKC SQSNIISTD EIIECLQNAK IQDIENWKGN NLAIIKGLIR TYNEEKNRLV EFFEDNCVNS LYLVEKLKEI INSGSITVGK SVTSKFIRNN HPLTVETYL KTKLYYRNNV TVLKSKKVSE ELYDLVKQFH NMMEIDLDSV MNLGKGTEGK KHTFLQMLEF VMSKAKNVTG S VDFLVSVF EKMQRTKTDR EIYLMSMKVK MMLYFIEHTF KHVAQSDPSE AISISGDNKI RALSTLSLDT ITSYNDILNK NS KKSRLAF LSADQSKWSA SDLTYKYVLA IILNPILTTG EASLMIECIL MYVKLKKVCI PTDIFLNLRK AQGTFGQNET AIG LLTKGL TTNTYPVSMN WLQGNLNYLS SVYHSCAMKA YHKTLECYKD CDFQTRWIVH SDDNATSLIA SGEVDKMLTD FSSS SLPEM LFRSIEAHFK SFCITLNPKK SYASSSEVEF ISERIVNGAI IPLYCRHLAN CCTESSHISY FDDLMSLSIH VTMLL RKGC PNEVIPFAYG AVQVQALSIY SMLPGEVNDS IRIFKKLGVS LKSNEIPTNM GGWLTSPIEP LSILGPSSND QIIYYN VIR DFLNKKSLEE VKDSVSSSSY LQMRFRELKG KYEKGTLEEK DKKMIFLINL FEKASVSEDS DVLTIGMKFQ TMLTQII KL PNFINENALN KMSSYKDFSK LYPNLKKNED LYKSTAKNLK IDEDAILEED ELYEKIASSL EMESVHDIMI KNPETILI A PLNDRDFLLS QLFMYTSPSK RNQLSNQSTA KLALDRVLRS KARTFVDISS TEKMTYEENM EKKILEMLKF DLDSYCSFK TCVNLVIKDV NFSMLIPILD SAYPCESRKR DNYNFRWFQT EKWIPVVEGS PGLVVMHAVY GSNYIENLGL KNIPLTDDSI NVLTSTFGT GLIMEDVKSL VKGKDSFETE AFSNSNECQR LVKACNYMIA AQNRLLAINT CFTRKSFPFY SKFNLGRGFI S NTLALL UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #2: RIBAVIRIN TRIPHOSPHATE
| Macromolecule | Name: RIBAVIRIN TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: RTP |
|---|---|
| Molecular weight | Theoretical: 484.144 Da |
| Chemical component information | 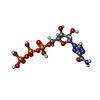 ChemComp-RTP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 144209 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: RANDOM ASSIGNMENT |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































