[English] 日本語
 Yorodumi
Yorodumi- EMDB-37252: Structure of tomato spotted wilt virus L protein binding to Ribavirin -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of tomato spotted wilt virus L protein binding to Ribavirin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | tomato spotted wilt virus / L protein / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA-directed RNA polymerase / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription Similarity search - Function | |||||||||
| Biological species |  Orthotospovirus tomatomaculae Orthotospovirus tomatomaculae | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Cao L / Wang X | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2025 Journal: Nat Plants / Year: 2025Title: Structural basis for the activation of plant bunyavirus replication machinery and its dual-targeted inhibition by ribavirin. Authors: Jia Li / Lei Cao / Yaqian Zhao / Jinghan Shen / Lei Wang / Mingfeng Feng / Min Zhu / Yonghao Ye / Richard Kormelink / Xiaorong Tao / Xiangxi Wang /   Abstract: Despite the discovery of plant viruses as a new class of pathogens over a century ago, the structure of plant virus replication machinery and antiviral pesticide remains lacking. Here we report five ...Despite the discovery of plant viruses as a new class of pathogens over a century ago, the structure of plant virus replication machinery and antiviral pesticide remains lacking. Here we report five cryogenic electron microscopy structures of a ~330-kDa RNA-dependent RNA polymerase (RdRp) from a devastating plant bunyavirus, tomato spotted wilt orthotospovirus (TSWV), including the apo, viral-RNA-bound, base analogue ribavirin-bound and ribavirin-triphosphate-bound states. They reveal that a flexible loop of RdRp's motif F functions as 'sensor' to perceive viral RNA and further acts as an 'adaptor' to promote the formation of a complete catalytic centre. A ten-base RNA 'hook' structure is sufficient to trigger major conformational changes and activate RdRp. Chemical screening showed that ribavirin is effective against TSWV, and structural data revealed that ribavirin disrupts both hook-binding and catalytic core formation, locking polymerase in its inactive state. This work provides structural insights into the mechanisms of plant bunyavirus RdRp activation and its dual-targeted site inhibition, facilitating the development of pesticides against plant viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37252.map.gz emd_37252.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37252-v30.xml emd-37252-v30.xml emd-37252.xml emd-37252.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37252.png emd_37252.png | 78.4 KB | ||
| Filedesc metadata |  emd-37252.cif.gz emd-37252.cif.gz | 7.1 KB | ||
| Others |  emd_37252_half_map_1.map.gz emd_37252_half_map_1.map.gz emd_37252_half_map_2.map.gz emd_37252_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37252 http://ftp.pdbj.org/pub/emdb/structures/EMD-37252 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37252 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37252 | HTTPS FTP |
-Related structure data
| Related structure data |  8ki6MC  8ki7C  8ki8C  8ki9C  8kiaC  9j8vC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_37252.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37252.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Structure of tomato spotted wilt virus L protein binding to Ribavirin
| Entire | Name: Structure of tomato spotted wilt virus L protein binding to Ribavirin |
|---|---|
| Components |
|
-Supramolecule #1: Structure of tomato spotted wilt virus L protein binding to Ribavirin
| Supramolecule | Name: Structure of tomato spotted wilt virus L protein binding to Ribavirin type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Orthotospovirus tomatomaculae Orthotospovirus tomatomaculae |
-Macromolecule #1: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Orthotospovirus tomatomaculae Orthotospovirus tomatomaculae |
| Molecular weight | Theoretical: 204.262906 KDa |
| Recombinant expression | Organism:  Spodoptera frugiperda multiple nucleopolyhedrovirus Spodoptera frugiperda multiple nucleopolyhedrovirus |
| Sequence | String: VFFSHWTSKY KERNPTEIAY SEDIERIIDS LVTDEITKEE IIHFLFGNFC FHIETMNDQH IADKFKGYQS SCINLKIEPK VDLADLKDH LIQKQQIWES LYGKHLEKIM LRIREKKKKE KEIPDITTAF NQNAAEYEEK YPNCFTNDLS ETKTNFSMTW S PSFEKIEL ...String: VFFSHWTSKY KERNPTEIAY SEDIERIIDS LVTDEITKEE IIHFLFGNFC FHIETMNDQH IADKFKGYQS SCINLKIEPK VDLADLKDH LIQKQQIWES LYGKHLEKIM LRIREKKKKE KEIPDITTAF NQNAAEYEEK YPNCFTNDLS ETKTNFSMTW S PSFEKIEL SSEVDYNNAI INKFRESFKS SSRVIYNSPY SSINNQTNKA RDITNLVRLC LTELSCDTTK MEKQELEDEI DI NTGSIKV ERTKKSKEWN KQGSCLTRNK NEFCMKETGR ENKTIYFKGL AVMNIGMSSK KRILKKEEIK ERISKGLEYD TSE RQADPN DDYSSIDMSS LTHMKKLIRH DNEDSLSWCE RIKDSLFVLH NGDIREEGKI TSVYNNYAKN PECLYIQDSV LKTE LETCK KINKLCNDLA IYHYSEDMMQ FSKGLMVADR YMTKESFKIL TTANTSMMLL AFKGDGMNTG GSGVPYIALH IVDED MSDQ FNICYTKEIY SYFRNGSNYI YIMRPQRLNQ VRLLSLFKTP SKVPVCFAQF SKKANEMEKW LKNKDIEKVN VFSMTM TVK QILINIVFSS VMIGTVTKLS RMGIFDFMRY AGFLPLSDYS NIKEYIRDKF DPDITNVADI YFVNGIKKLL FRMEDLN LS TNAKPVVVDH ENDIIGGITD LNIKCPITGS TLLTLEDLYN NVYLAIYMMP KSLHNHVHNL TSLLNVPAEW ELKFRKEL G FNIFEDIYPK KAMFDDKDLF SINGALNVKA LSDYYLGNIE NVGLMRSEIE NKEDFLSPCY KISTLKSSKK CSQSNIIST DEIIECLQNA KIQDIENWKG NNLAIIKGLI RTYNEEKNRL VEFFEDNCVN SLYLVEKLKE IINSGSITVG KSVTSKFIRN NHPLTVETY LKTKLYYRNN VTVLKSKKVS EELYDLVKQF HNMMEIDLDS VMNLGKGTEG KKHTFLQMLE FVMSKAKNVT G SVDFLVSV FEKMQRTKTD REIYLMSMKV KMMLYFIEHT FKHVAQSDPS EAISISGDNK IRALSTLSLD TITSYNDILN KN SKKSRLA FLSADQSKWS ASDLTYKYVL AIILNPILTT GEASLMIECI LMYVKLKKVC IPTDIFLNLR KAQGTFGQNE TAI GLLTKG LTTNTYPVSM NWLQGNLNYL SSVYHSCAMK AYHKTLECYK DCDFQTRWIV HSDDNATSLI ASGEVDKMLT DFSS SSLPE MLFRSIEAHF KSFCITLNPK KSYASSSEVE FISERIVNGA IIPLYCRHLA NCCTESSHIS YFDDLMSLSI HVTML LRKG CPNEVIPFAY GAVQVQALSI YSMLPGEVND SIRIFKKLGV SLKSNEIPTN MGGWLTSPIE PLSILGPSSN DQIIYY NVI RDFLNKKSLE EVKDSVSSSS YLQMRFRELK GKYEKGTLEE KDKKMIFLIN LFEKASVSED SDVLTIGMKF QTMLTQI IK LPNFINENAL NKMSSYKDFS KLYPNLKKNE DLYKSTKNLK IDEDAILEED ELYEKIASSL EMESVHDIMI KNPETILI A PLNDRDFLLS QLFMYTSPSK RNQLSNQSTE KLALDRVLRS KARTFVDISS TVKMTYEENM EKKILEMLKF DLDSYCSFK TCVNLVIKDV NFSMLIPILD SAYPCESRKR DNYNFRWFQT EKWIPVVEGS PGLVVMHAVY GSNYIENLGL KNIPLTDDSI NVLTSTFGT GLIMEDVKSL VKGKDSFETE AFSNSNECQR LVKACNYMIA AQNRLLAINT CFTRKSFPFY SKFNLGRGFI S NTLALLST IYSKEES UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #2: 1-(beta-D-ribofuranosyl)-1H-1,2,4-triazole-3-carboxamide
| Macromolecule | Name: 1-(beta-D-ribofuranosyl)-1H-1,2,4-triazole-3-carboxamide type: ligand / ID: 2 / Number of copies: 2 / Formula: RBV |
|---|---|
| Molecular weight | Theoretical: 244.205 Da |
| Chemical component information | 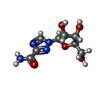 ChemComp-RBV: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















