+ Open data
Open data
- Basic information
Basic information
| Entry | 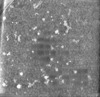 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron tomogram of ATG2A and small unilamellar vesicles | |||||||||
 Map data Map data | Cryo-electron tomogram of ATG2A and small unilamellar vesicles (denoised using IsoNet) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | autophagy / intracellular trafficking / membranes / vesicles / LIPID TRANSPORT | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Dahmane S / Wang N / Stjepanovic G / Carlson LA | |||||||||
| Funding support |  Sweden, 2 items Sweden, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2025 Journal: Nat Struct Mol Biol / Year: 2025Title: Structural basis for lipid transfer by the ATG2A-ATG9A complex. Authors: Yang Wang / Selma Dahmane / Rujuan Ti / Xinyi Mai / Lizhe Zhu / Lars-Anders Carlson / Goran Stjepanovic /   Abstract: Autophagy is characterized by the formation of double-membrane vesicles called autophagosomes. Autophagy-related proteins (ATGs) 2A and 9A have an essential role in autophagy by mediating lipid ...Autophagy is characterized by the formation of double-membrane vesicles called autophagosomes. Autophagy-related proteins (ATGs) 2A and 9A have an essential role in autophagy by mediating lipid transfer and re-equilibration between membranes for autophagosome formation. Here we report the cryo-electron microscopy structures of human ATG2A in complex with WD-repeat protein interacting with phosphoinositides 4 (WIPI4) at 3.2 Å and the ATG2A-WIPI4-ATG9A complex at 7 Å global resolution. On the basis of molecular dynamics simulations, we propose a mechanism of lipid extraction from the donor membranes. Our analysis revealed 3:1 stoichiometry of the ATG9A-ATG2A complex, directly aligning the ATG9A lateral pore with ATG2A lipid transfer cavity, and an interaction of the ATG9A trimer with both the N-terminal and the C-terminal tip of rod-shaped ATG2A. Cryo-electron tomography of ATG2A liposome-binding states showed that ATG2A tethers lipid vesicles at different orientations. In summary, this study provides a molecular basis for the growth of the phagophore membrane and lends structural insights into spatially coupled lipid transport and re-equilibration during autophagosome formation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50662.map.gz emd_50662.map.gz | 200.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50662-v30.xml emd-50662-v30.xml emd-50662.xml emd-50662.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_50662.png emd_50662.png | 202.6 KB | ||
| Filedesc metadata |  emd-50662.cif.gz emd-50662.cif.gz | 3.7 KB | ||
| Others |  emd_50662_additional_1.map.gz emd_50662_additional_1.map.gz | 206.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50662 http://ftp.pdbj.org/pub/emdb/structures/EMD-50662 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50662 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50662 | HTTPS FTP |
-Validation report
| Summary document |  emd_50662_validation.pdf.gz emd_50662_validation.pdf.gz | 344.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_50662_full_validation.pdf.gz emd_50662_full_validation.pdf.gz | 344.1 KB | Display | |
| Data in XML |  emd_50662_validation.xml.gz emd_50662_validation.xml.gz | 4.7 KB | Display | |
| Data in CIF |  emd_50662_validation.cif.gz emd_50662_validation.cif.gz | 5.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50662 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50662 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50662 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50662 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_50662.map.gz / Format: CCP4 / Size: 223.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50662.map.gz / Format: CCP4 / Size: 223.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-electron tomogram of ATG2A and small unilamellar vesicles (denoised using IsoNet) | ||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 11.12 Å | ||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: un-denoised map
| File | emd_50662_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | un-denoised map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ATG2A and small unilamellar vesicles
| Entire | Name: ATG2A and small unilamellar vesicles |
|---|---|
| Components |
|
-Supramolecule #1: ATG2A and small unilamellar vesicles
| Supramolecule | Name: ATG2A and small unilamellar vesicles / type: complex / ID: 1 / Parent: 0 / Details: Purified human protein mixed with vesicles (SUVs). |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
| Sectioning | Other: NO SECTIONING |
| Fiducial marker | Manufacturer: CMC-Utrecht / Diameter: 10 nm |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 2.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 3.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Number images used: 41 |
|---|
 Movie
Movie Controller
Controller












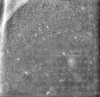
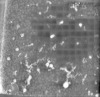





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)
























