+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-3284 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of BK polyomavirus VP1 virus-like particle | ||||||||||||
 マップデータ マップデータ | Reconstruction of BK VP1 VLP (sharpened/masked) | ||||||||||||
 試料 試料 |
| ||||||||||||
 キーワード キーワード | BKPyV / BK / polyomavirus / virus-like particle / VLP | ||||||||||||
| 生物種 | BK polyomavirus VP1 VLP | ||||||||||||
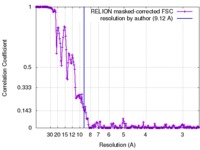
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 9.12 Å | ||||||||||||
 データ登録者 データ登録者 | Hurdiss DL / Morgan EL / Thompson RF / Prescott EL / Panou MM / Macdonald A / Ranson NA | ||||||||||||
 引用 引用 |  ジャーナル: Structure / 年: 2016 ジャーナル: Structure / 年: 2016タイトル: New Structural Insights into the Genome and Minor Capsid Proteins of BK Polyomavirus using Cryo-Electron Microscopy. 著者: Daniel L Hurdiss / Ethan L Morgan / Rebecca F Thompson / Emma L Prescott / Margarita M Panou / Andrew Macdonald / Neil A Ranson /  要旨: BK polyomavirus is the causative agent of several diseases in transplant patients and the immunosuppressed. In order to better understand the structure and life cycle of BK, we produced infectious ...BK polyomavirus is the causative agent of several diseases in transplant patients and the immunosuppressed. In order to better understand the structure and life cycle of BK, we produced infectious virions and VP1-only virus-like particles in cell culture, and determined their three-dimensional structures using cryo-electron microscopy (EM) and single-particle image processing. The resulting 7.6-Å resolution structure of BK and 9.1-Å resolution of the virus-like particles are the highest-resolution cryo-EM structures of any polyomavirus. These structures confirm that the architecture of the major structural protein components of these human polyomaviruses are similar to previous structures from other hosts, but give new insight into the location and role of the enigmatic minor structural proteins, VP2 and VP3. We also observe two shells of electron density, which we attribute to a structurally ordered part of the viral genome, and discrete contacts between this density and both VP1 and the minor capsid proteins. | ||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_3284.map.gz emd_3284.map.gz | 104.4 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-3284-v30.xml emd-3284-v30.xml emd-3284.xml emd-3284.xml | 8.8 KB 8.8 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_3284_fsc.xml emd_3284_fsc.xml | 17 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_3284.tif emd_3284.tif | 2.2 MB | ||
| その他 |  emd_3284_additional_1.map.gz emd_3284_additional_1.map.gz | 374.3 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3284 http://ftp.pdbj.org/pub/emdb/structures/EMD-3284 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3284 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3284 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_3284_validation.pdf.gz emd_3284_validation.pdf.gz | 311.2 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_3284_full_validation.pdf.gz emd_3284_full_validation.pdf.gz | 310.3 KB | 表示 | |
| XML形式データ |  emd_3284_validation.xml.gz emd_3284_validation.xml.gz | 15.4 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3284 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3284 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3284 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3284 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_3284.map.gz / 形式: CCP4 / 大きさ: 465.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_3284.map.gz / 形式: CCP4 / 大きさ: 465.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Reconstruction of BK VP1 VLP (sharpened/masked) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
-添付マップデータ: emd 3284 additional 1.map
| ファイル | emd_3284_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : BK polyomavirus VP1 VLP
| 全体 | 名称: BK polyomavirus VP1 VLP |
|---|---|
| 要素 |
|
-超分子 #1000: BK polyomavirus VP1 VLP
| 超分子 | 名称: BK polyomavirus VP1 VLP / タイプ: sample / ID: 1000 / 集合状態: Icosohedral / Number unique components: 1 |
|---|
-超分子 #1: BK polyomavirus VP1 VLP
| 超分子 | 名称: BK polyomavirus VP1 VLP / タイプ: virus / ID: 1 / 生物種: BK polyomavirus VP1 VLP / Sci species strain: BKV-Ia / ウイルスタイプ: VIRUS-LIKE PARTICLE / ウイルス・単離状態: STRAIN / ウイルス・エンベロープ: No / ウイルス・中空状態: No |
|---|---|
| 宿主 | 生物種:  Homo sapiens (ヒト) / 別称: VERTEBRATES Homo sapiens (ヒト) / 別称: VERTEBRATES |
| Host system | 組換細胞: HEK293TT / 組換プラスミド: pIaw |
| ウイルス殻 | Shell ID: 1 / 直径: 498 Å |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.9 / 詳細: 10mM HEPES pH 7.9, 50mM CaCl2, 1mM MgCl2, 5mM KCl |
|---|---|
| グリッド | 詳細: Quantifoil R2/1 EM grids |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / 装置: FEI VITROBOT MARK IV / 手法: 6.5 seconds blot before plunging |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI POLARA 300 |
|---|---|
| 日付 | 2015年5月13日 |
| 撮影 | カテゴリ: CCD フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 実像数: 170 / 平均電子線量: 40 e/Å2 詳細: 4 e-/A2/s, a 4 frames per second frame rate, and a 10 s exposure |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 6.136 µm / 最小 デフォーカス(公称値): 0.526 µm / 倍率(公称値): 19000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER |
| 実験機器 |  モデル: Tecnai Polara / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)