[English] 日本語
 Yorodumi
Yorodumi- EMDB-3198: Cryo-EM structure of the E. coli replicative DNA polymerase compl... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3198 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon, and tau subunits) | |||||||||
 Map data Map data | E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon, and tau subunits) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA replication / DNA polymerase III alpha / DNA polymerase III beta / DNA polymerase III epsilon / DNA polymerase III tau | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA polymerase III, core complex / DNA polymerase III, clamp loader complex / DNA clamp loader activity / DNA polymerase III complex / DNA replication proofreading / replisome / lagging strand elongation / DNA strand elongation involved in DNA replication / exonuclease activity / leading strand elongation ...DNA polymerase III, core complex / DNA polymerase III, clamp loader complex / DNA clamp loader activity / DNA polymerase III complex / DNA replication proofreading / replisome / lagging strand elongation / DNA strand elongation involved in DNA replication / exonuclease activity / leading strand elongation / DNA polymerase processivity factor activity / 3'-5' exonuclease activity / ribonucleoside triphosphate phosphatase activity / DNA-templated DNA replication / DNA replication / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / ribosome / structural constituent of ribosome / ribonucleoprotein complex / translation / viral translational frameshifting / ATP hydrolysis activity / DNA binding / ATP binding / identical protein binding / metal ion binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
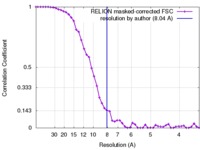
| Method | single particle reconstruction / cryo EM / Resolution: 8.04 Å | |||||||||
 Authors Authors | Fernandez-Leiro R / Conrad J / Scheres SHW / Lamers MH | |||||||||
 Citation Citation |  Journal: Elife / Year: 2015 Journal: Elife / Year: 2015Title: cryo-EM structures of the replicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease and . Authors: Rafael Fernandez-Leiro / Julian Conrad / Sjors Hw Scheres / Meindert H Lamers /  Abstract: The replicative DNA polymerase PolIIIα from is a uniquely fast and processive enzyme. For its activity it relies on the DNA sliding clamp β, the proofreading exonuclease ε and the C-terminal ...The replicative DNA polymerase PolIIIα from is a uniquely fast and processive enzyme. For its activity it relies on the DNA sliding clamp β, the proofreading exonuclease ε and the C-terminal domain of the clamp loader subunit τ. Due to the dynamic nature of the four-protein complex it has long been refractory to structural characterization. Here we present the 8 Å resolution cryo-electron microscopy structures of DNA-bound and DNA-free states of the PolIII-clamp-exonuclease-τ complex. The structures show how the polymerase is tethered to the DNA through multiple contacts with the clamp and exonuclease. A novel contact between the polymerase and clamp is made in the DNA bound state, facilitated by a large movement of the polymerase tail domain and τ. These structures provide crucial insights into the organization of the catalytic core of the replisome and form an important step towards determining the structure of the complete holoenzyme. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3198.map.gz emd_3198.map.gz | 7.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3198-v30.xml emd-3198-v30.xml emd-3198.xml emd-3198.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3198_fsc.xml emd_3198_fsc.xml | 4.6 KB | Display |  FSC data file FSC data file |
| Images |  EMD-3198.png EMD-3198.png emd_3198.png emd_3198.png | 277.2 KB 277.2 KB | ||
| Masks |  emd_3198_msk_1.map emd_3198_msk_1.map | 8 MB |  Mask map Mask map | |
| Others |  emd_3198_half_map_1.map.gz emd_3198_half_map_1.map.gz emd_3198_half_map_2.map.gz emd_3198_half_map_2.map.gz | 6 MB 6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3198 http://ftp.pdbj.org/pub/emdb/structures/EMD-3198 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3198 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3198 | HTTPS FTP |
-Validation report
| Summary document |  emd_3198_validation.pdf.gz emd_3198_validation.pdf.gz | 315 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3198_full_validation.pdf.gz emd_3198_full_validation.pdf.gz | 314.2 KB | Display | |
| Data in XML |  emd_3198_validation.xml.gz emd_3198_validation.xml.gz | 7.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3198 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3198 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3198 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3198 | HTTPS FTP |
-Related structure data
| Related structure data |  5fkvMC  3201C  3202C  5fkuC  5fkwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3198.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3198.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon, and tau subunits) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.76 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Segmentation: relion automask for reconstruction
| Annotation | relion automask for reconstruction | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| File |  emd_3198_msk_1.map emd_3198_msk_1.map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Supplemental map: emd 3198 half map 1.map
| File | emd_3198_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Supplemental map: emd 3198 half map 2.map
| File | emd_3198_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : E. coli replicative DNA polymerase complex bound to DNA (DNA poly...
| Entire | Name: E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon, and tau subunits) |
|---|---|
| Components |
|
-Supramolecule #1000: E. coli replicative DNA polymerase complex bound to DNA (DNA poly...
| Supramolecule | Name: E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon, and tau subunits) type: sample / ID: 1000 Details: Individual proteins purified individually and the complex was later assembled in vitro and purified over gel filtration Oligomeric state: 1 DNA polymerase III alpha : 2 DNA polymerase III beta: 1 DNA polymerase III epsilon : 1 DNA polymerase III tau : dsDNA Number unique components: 5 |
|---|---|
| Molecular weight | Theoretical: 256 KDa |
-Macromolecule #1: DNA polymerase III alpha
| Macromolecule | Name: DNA polymerase III alpha / type: protein_or_peptide / ID: 1 / Name.synonym: dnaE Details: amino acid residues 920-924 of E. coli PolIII alpha were changed by site directed mutagenesis from QADMF to QLDLF Number of copies: 1 / Oligomeric state: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 132 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: DNA polymerase III subunit alpha GO: DNA replication, DNA binding, DNA-directed DNA polymerase activity InterPro: DNA polymerase III, alpha subunit |
-Macromolecule #2: DNA polymerase III beta
| Macromolecule | Name: DNA polymerase III beta / type: protein_or_peptide / ID: 2 / Name.synonym: DNA clamp / Number of copies: 1 / Oligomeric state: 2 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 81 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Large ribosomal subunit protein bL34 / GO: DNA strand elongation involved in DNA replication / InterPro: DNA polymerase III, beta sliding clamp |
-Macromolecule #3: DNA polymerase III epsilon
| Macromolecule | Name: DNA polymerase III epsilon / type: protein_or_peptide / ID: 3 / Name.synonym: dnaQ Details: amino acid residues 182-187 of E. coli PolIII epsilon were changed by site directed mutagenesis from QTSMAF to QLSLPL Number of copies: 1 / Oligomeric state: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.3762 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: DNA polymerase III subunit epsilon / GO: exonuclease activity / InterPro: DNA polymerase 3, epsilon subunit |
-Macromolecule #4: DNA polymerase III tau
| Macromolecule | Name: DNA polymerase III tau / type: protein_or_peptide / ID: 4 / Name.synonym: dnaX / Details: polymerase-binding domain of tau: residues 500-643 / Number of copies: 1 / Oligomeric state: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.264 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: DNA polymerase III subunit tau / GO: exonuclease activity / InterPro: DNA polymerase III, tau subunit, domain V |
-Macromolecule #5: primer-template duplex DNA
| Macromolecule | Name: primer-template duplex DNA / type: dna / ID: 5 / Name.synonym: dsDNA Details: template strans has a 4 nucleotide overhang, sequence as follows: TCAGGAGTCCTTCGTCCTAGTACTACTCC Classification: DNA / Structure: DOUBLE HELIX / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism: unidentified (others) |
| Molecular weight | Theoretical: 15.5 KDa |
| Sequence | String: GGAGTAGTAC TAGGACGAAG GACTC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 25 mM Hepes pH 7.5, 50 mM Potassium Glutamate, 3 mM Magnesium Acetate, 2 mM DTT |
| Grid | Details: glow-discharged holey carbon cryo-EM grids (Quantifoil Cu R1.2/1.3 400 mesh) |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 110 K / Instrument: FEI VITROBOT MARK III Method: Prior to sample preparation 0.1 volumes of 0.05% Tween 20 were added to the sample 3 microliters were pipetted onto the grid and blotted for 4 seconds |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 80 K / Max: 90 K / Average: 85 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 64,000 times magnification |
| Date | May 12, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Digitization - Sampling interval: 5 µm / Number real images: 1350 / Average electron dose: 40 e/Å2 / Details: 20 frames/micrograph / Bits/pixel: 32 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 28409 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)