+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30327 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mycobacterium smegmatis SugABC complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC transporter / Lipoprotein / Mycobacteria / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcarbohydrate transport / ABC-type transporter activity / ATP-binding cassette (ABC) transporter complex / transmembrane transport / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) / Mycolicibacterium smegmatis MC2 155 (bacteria) /  Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.41 Å | |||||||||
 Authors Authors | Liu F / Liang J | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2020 Journal: Sci Adv / Year: 2020Title: Structural basis of trehalose recycling by the ABC transporter LpqY-SugABC. Authors: Fengjiang Liu / Jingxi Liang / Bing Zhang / Yan Gao / Xiuna Yang / Tianyu Hu / Haitao Yang / Wenqing Xu / Luke W Guddat / Zihe Rao /   Abstract: In bacteria, adenosine 5'-triphosphate (ATP)-binding cassette (ABC) importers are essential for the uptake of nutrients including the nonreducing disaccharide trehalose, a metabolite that is crucial ...In bacteria, adenosine 5'-triphosphate (ATP)-binding cassette (ABC) importers are essential for the uptake of nutrients including the nonreducing disaccharide trehalose, a metabolite that is crucial for the survival and virulence of several human pathogens including SugABC is an ABC transporter that translocates trehalose from the periplasmic lipoprotein LpqY into the cytoplasm of mycobacteria. Here, we report four high-resolution cryo-electron microscopy structures of the mycobacterial LpqY-SugABC complex to reveal how it binds and passes trehalose through the membrane to the cytoplasm. A unique feature observed in this system is the initial mode of capture of the trehalose at the LpqY interface. Uptake is achieved by a pivotal rotation of LpqY relative to SugABC, moving from an open and accessible conformation to a clamped conformation upon trehalose binding. These findings enrich our understanding as to how ABC transporters facilitate substrate transport across the membrane in Gram-positive bacteria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30327.map.gz emd_30327.map.gz | 200.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30327-v30.xml emd-30327-v30.xml emd-30327.xml emd-30327.xml | 12.4 KB 12.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30327.png emd_30327.png | 52.8 KB | ||
| Filedesc metadata |  emd-30327.cif.gz emd-30327.cif.gz | 5.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30327 http://ftp.pdbj.org/pub/emdb/structures/EMD-30327 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30327 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30327 | HTTPS FTP |
-Validation report
| Summary document |  emd_30327_validation.pdf.gz emd_30327_validation.pdf.gz | 546.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30327_full_validation.pdf.gz emd_30327_full_validation.pdf.gz | 545.9 KB | Display | |
| Data in XML |  emd_30327_validation.xml.gz emd_30327_validation.xml.gz | 7 KB | Display | |
| Data in CIF |  emd_30327_validation.cif.gz emd_30327_validation.cif.gz | 8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30327 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30327 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30327 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30327 | HTTPS FTP |
-Related structure data
| Related structure data |  7cadMC  7caeC  7cafC  7cagC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30327.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30327.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
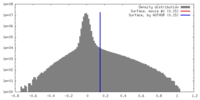
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Mycobacterium smegmatis SugABC complex
| Entire | Name: Mycobacterium smegmatis SugABC complex |
|---|---|
| Components |
|
-Supramolecule #1: Mycobacterium smegmatis SugABC complex
| Supramolecule | Name: Mycobacterium smegmatis SugABC complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
-Macromolecule #1: ABC transporter, ATP-binding protein SugC
| Macromolecule | Name: ABC transporter, ATP-binding protein SugC / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 43.703609 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MAEIVLDRVT KSYPDGAGGV RAAVKEFSMT IADGEFIILV GPSGCGKSTT LNMIAGLEEI TSGELRIGGE RVNEKAPKDR DIAMVFQSY ALYPHMTVRQ NIAFPLTLAK VPKAEIAAKV EETAKILDLS ELLDRKPGQL SGGQRQRVAM GRAIVRSPKA F LMDEPLSN ...String: MAEIVLDRVT KSYPDGAGGV RAAVKEFSMT IADGEFIILV GPSGCGKSTT LNMIAGLEEI TSGELRIGGE RVNEKAPKDR DIAMVFQSY ALYPHMTVRQ NIAFPLTLAK VPKAEIAAKV EETAKILDLS ELLDRKPGQL SGGQRQRVAM GRAIVRSPKA F LMDEPLSN LDAKLRVQMR AEISRLQDRL GTTTVYVTHD QTEAMTLGDR VVVMLAGEVQ QIGTPDELYS SPANLFVAGF IG SPAMNFF PATRTDVGVR LPFGEVTLTP HMLDLLDKQA RPENIIVGIR PEHIEDSALL DGYARIRALT FSVRADIVES LGA DKYVHF TTEGAGAESA QLAELAADSG AGTNQFIARV SADSRVRTGE QIELAIDTTK LSIFDAATGL NLTRDITPTD PTEA AGPDA G UniProtKB: ABC transporter, ATP-binding protein SugC |
-Macromolecule #2: ABC sugar transporter, permease component
| Macromolecule | Name: ABC sugar transporter, permease component / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 32.73925 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MTAAVTPSAS AVASDDKKSE RRLAFWLIAP AVLLMLAVTA YPIGYAVWLS LQRYNLAEPH DTEFIGLANY VTVLTDGYWW TAFAVTLGI TVVSVAIEFA LGLALALVMH RTIFGKGAVR TAILIPYGIV TVAASYSWYY AWTPGTGYLA NLLPEGSAPL T DQLPSLAI ...String: MTAAVTPSAS AVASDDKKSE RRLAFWLIAP AVLLMLAVTA YPIGYAVWLS LQRYNLAEPH DTEFIGLANY VTVLTDGYWW TAFAVTLGI TVVSVAIEFA LGLALALVMH RTIFGKGAVR TAILIPYGIV TVAASYSWYY AWTPGTGYLA NLLPEGSAPL T DQLPSLAI VVLAEVWKTT PFMALLLLAG LALVPQDLLN AAQVDGAGPW KRLTKVILPM IKPAILVALL FRTLDAFRIF DN IYILTGG SNDTGSVSIL GYDNLFKAFN VGLGSAISVL IFLSVAIIAF IYIKIFGAAA PGSDEEVR UniProtKB: ABC sugar transporter, permease component |
-Macromolecule #3: ABC transporter, permease protein SugB
| Macromolecule | Name: ABC transporter, permease protein SugB / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 29.839441 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MADRVDARRA TWWSVVNILV IVYALIPVLW ILSLSLKPTS SVKDGKLIPT EITFANYKAI FSGDAFTSAL FNSIGIGLIT TIIAVVIGG MAAYAVARLQ FPGKQLLIGV ALLIAMFPHI SLVTPIFNMW RGIGLFDTWP GLIIPYITFA LPLAIYTLSA F FREIPWDL ...String: MADRVDARRA TWWSVVNILV IVYALIPVLW ILSLSLKPTS SVKDGKLIPT EITFANYKAI FSGDAFTSAL FNSIGIGLIT TIIAVVIGG MAAYAVARLQ FPGKQLLIGV ALLIAMFPHI SLVTPIFNMW RGIGLFDTWP GLIIPYITFA LPLAIYTLSA F FREIPWDL EKAAKMDGAT PAQAFRKVIA PLAAPGIVTA AILVFIFAWN DLLLALSLTA TQRAITAPVA IANFTGSSQF EE PTGSIAA GAMVITIPII IFVLIFQRRI VAGLTSGAVK G UniProtKB: ABC transporter, permease protein SugB |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.41 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 197191 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)