+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2982 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
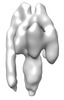
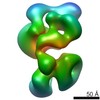
| Title | Sub-tomogram average of a mammalian F-type ATP synthase monomer | |||||||||
 Map data Map data | Sub-tomogram average of bovine F-type ATP synthase in a 2D crystal | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | F-type ATP synthase / Mitochondria Bovine heart | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / negative staining / Resolution: 24.0 Å | |||||||||
 Authors Authors | Jiko C / Davies KM / Shinzawa-Itoh K / Tani K / Maeda S / Mills DJ / Tsukihara T / Fujiyoshi Y / Kuehlbrandt W / Gerle C | |||||||||
 Citation Citation |  Journal: Elife / Year: 2015 Journal: Elife / Year: 2015Title: Bovine F1Fo ATP synthase monomers bend the lipid bilayer in 2D membrane crystals. Authors: Chimari Jiko / Karen M Davies / Kyoko Shinzawa-Itoh / Kazutoshi Tani / Shintaro Maeda / Deryck J Mills / Tomitake Tsukihara / Yoshinori Fujiyoshi / Werner Kühlbrandt / Christoph Gerle /   Abstract: We have used a combination of electron cryo-tomography, subtomogram averaging, and electron crystallographic image processing to analyse the structure of intact bovine F(1)F(o) ATP synthase in 2D ...We have used a combination of electron cryo-tomography, subtomogram averaging, and electron crystallographic image processing to analyse the structure of intact bovine F(1)F(o) ATP synthase in 2D membrane crystals. ATPase assays and mass spectrometry analysis of the 2D crystals confirmed that the enzyme complex was complete and active. The structure of the matrix-exposed region was determined at 24 Å resolution by subtomogram averaging and repositioned into the tomographic volume to reveal the crystal packing. F(1)F(o) ATP synthase complexes are inclined by 16° relative to the crystal plane, resulting in a zigzag topology of the membrane and indicating that monomeric bovine heart F(1)F(o) ATP synthase by itself is sufficient to deform lipid bilayers. This local membrane curvature is likely to be instrumental in the formation of ATP synthase dimers and dimer rows, and thus for the shaping of mitochondrial cristae. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2982.map.gz emd_2982.map.gz | 339 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2982-v30.xml emd-2982-v30.xml emd-2982.xml emd-2982.xml | 11.3 KB 11.3 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2982.png EMD-2982.png | 90.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2982 http://ftp.pdbj.org/pub/emdb/structures/EMD-2982 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2982 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2982 | HTTPS FTP |
-Validation report
| Summary document |  emd_2982_validation.pdf.gz emd_2982_validation.pdf.gz | 192.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2982_full_validation.pdf.gz emd_2982_full_validation.pdf.gz | 192 KB | Display | |
| Data in XML |  emd_2982_validation.xml.gz emd_2982_validation.xml.gz | 4.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2982 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2982 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2982 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2982 | HTTPS FTP |
-Related structure data
| Similar structure data | |
|---|---|
| EM raw data |  EMPIAR-10027 (Title: Sub-tomogram average of a mammalian F-type ATP synthase monomer EMPIAR-10027 (Title: Sub-tomogram average of a mammalian F-type ATP synthase monomerData size: 8.9 / Data #1: 2dxtal_A [class averages] / Data #2: 2dxtal_B [class averages]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2982.map.gz / Format: CCP4 / Size: 429.7 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2982.map.gz / Format: CCP4 / Size: 429.7 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sub-tomogram average of bovine F-type ATP synthase in a 2D crystal | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : 2D crystal of bovine F-type ATP synthase monomers
| Entire | Name: 2D crystal of bovine F-type ATP synthase monomers |
|---|---|
| Components |
|
-Supramolecule #1000: 2D crystal of bovine F-type ATP synthase monomers
| Supramolecule | Name: 2D crystal of bovine F-type ATP synthase monomers / type: sample / ID: 1000 Details: 2D Membrane crystals lipids for reconstitution: 1,2-dimyristoyl-sn-glycero-3-phosphocholine Oligomeric state: 1 / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 600 KDa |
-Macromolecule #1: F-type ATP synthase
| Macromolecule | Name: F-type ATP synthase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 600 KDa |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 8.2 Details: 40mM Tris-HCL pH8.2 100mM NaCl 0.02%(w/v)NaN3, 0.5mM ADP, 5mM MgCl2, 0.1mM DTT, 0.1mM EDTA |
| Staining | Type: NEGATIVE Details: Plunge frozen in liquid ethane using home-made freezing device |
| Grid | Details: Glow discharged R2/2, 300 copper mesh quantifoil grids |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 100 K / Instrument: HOMEMADE PLUNGER Method: 2D crystals were mixed 1:1 (v/v) with 6nm collodial gold particles. Three microliters of protein/gold sample was applied to R2/2 300 copper mesh quantifoil grids. Access liquid was removed by ...Method: 2D crystals were mixed 1:1 (v/v) with 6nm collodial gold particles. Three microliters of protein/gold sample was applied to R2/2 300 copper mesh quantifoil grids. Access liquid was removed by blotting for 3s with Whatman #4 paper before plunge-freezing in liquid ethane |
| Details | 10mg/ml ATP synthase were mixed with 1,2-dimyristoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids) using a lipid to protein ratio of 0.2. Detergent was removed by dialysis using 20 microliter Hampton dialysis buttons and 15kDa membrane cuttoff. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 80 K / Max: 100 K / Average: 90 K |
| Alignment procedure | Legacy - Astigmatism: Objection lend astigmation was corrected on K2 at magnification used for imaging. |
| Specialist optics | Energy filter - Name: quantum / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Details | tomographic tilt series |
| Date | Feb 1, 2013 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Number real images: 80 / Average electron dose: 60 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 14942 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 42000 |
| Sample stage | Specimen holder: Nitrogen cooled / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Tilt series - Axis1 - Min angle: -60 ° / Tilt series - Axis1 - Max angle: 60 ° |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Subtomograms were selected manually and averaged using PEET (Particle Estimation for Electron Tomography, Boulder) |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 24.0 Å / Resolution method: OTHER / Software - Name:  IMOD / Number subtomograms used: 2100 IMOD / Number subtomograms used: 2100 |
| CTF correction | Details: IMOD |
| Crystal parameters | Unit cell - A: 179.1 Å / Unit cell - B: 171.4 Å / Unit cell - γ: 94.9 ° / Plane group: P 1 |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















