[English] 日本語
 Yorodumi
Yorodumi- EMDB-3790: Structure of Tra1 subunit within the chromatin modifying complex SAGA -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3790 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
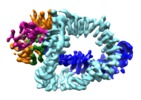
| Title | Structure of Tra1 subunit within the chromatin modifying complex SAGA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | yeast / Tra1 / SAGA / activator target / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationSAGA complex / NuA4 histone acetyltransferase complex / transferase activity / DNA repair / regulation of DNA-templated transcription / nucleus Similarity search - Function | |||||||||
| Biological species |  Komagataella pastoris (fungus) Komagataella pastoris (fungus) | |||||||||
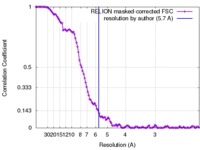
| Method | single particle reconstruction / cryo EM / Resolution: 5.7 Å | |||||||||
 Authors Authors | Sharov G / Voltz K | |||||||||
| Funding support |  France, 2 items France, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2017 Journal: Nat Commun / Year: 2017Title: Structure of the transcription activator target Tra1 within the chromatin modifying complex SAGA. Authors: Grigory Sharov / Karine Voltz / Alexandre Durand / Olga Kolesnikova / Gabor Papai / Alexander G Myasnikov / Annick Dejaegere / Adam Ben Shem / Patrick Schultz /    Abstract: The transcription co-activator complex SAGA is recruited to gene promoters by sequence-specific transcriptional activators and by chromatin modifications to promote pre-initiation complex formation. ...The transcription co-activator complex SAGA is recruited to gene promoters by sequence-specific transcriptional activators and by chromatin modifications to promote pre-initiation complex formation. The yeast Tra1 subunit is the major target of acidic activators such as Gal4, VP16, or Gcn4 but little is known about its structural organization. The 430 kDa Tra1 subunit and its human homolog the transformation/transcription domain-associated protein TRRAP are members of the phosphatidyl 3-kinase-related kinase (PIKK) family. Here, we present the cryo-EM structure of the entire SAGA complex where the major target of activator binding, the 430 kDa Tra1 protein, is resolved with an average resolution of 5.7 Å. The high content of alpha-helices in Tra1 enabled tracing of the majority of its main chain. Our results highlight the integration of Tra1 within the major epigenetic regulator SAGA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3790.map.gz emd_3790.map.gz | 7.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3790-v30.xml emd-3790-v30.xml emd-3790.xml emd-3790.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3790_fsc.xml emd_3790_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_3790.png emd_3790.png | 214.4 KB | ||
| Filedesc metadata |  emd-3790.cif.gz emd-3790.cif.gz | 8.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3790 http://ftp.pdbj.org/pub/emdb/structures/EMD-3790 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3790 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3790 | HTTPS FTP |
-Related structure data
| Related structure data |  5oejMC  3804C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
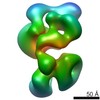
Map
| File |  Download / File: emd_3790.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3790.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Tra1 subunit of SAGA complex
| Entire | Name: Tra1 subunit of SAGA complex |
|---|---|
| Components |
|
-Supramolecule #1: Tra1 subunit of SAGA complex
| Supramolecule | Name: Tra1 subunit of SAGA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: Tra1 subunit within the chromatin modifying complex SAGA
| Macromolecule | Name: Tra1 subunit within the chromatin modifying complex SAGA type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Molecular weight | Theoretical: 438.055344 KDa |
| Sequence | String: MLHVVQLDDF ATRLKAAEDY QSKHSVLSEI CDSLETFNAA QDYEYFLKSL IPLFIDVLKE VPVSFVANSP ENKLRNITLE ILHRIPAND ALQAYSNEIV DTLMDLLKVE NELNGILCMK AITTLHKTFK ASLQEKVHPF IDIVIEIYSN IPQVVEEQFN G NQIDSKEN ...String: MLHVVQLDDF ATRLKAAEDY QSKHSVLSEI CDSLETFNAA QDYEYFLKSL IPLFIDVLKE VPVSFVANSP ENKLRNITLE ILHRIPAND ALQAYSNEIV DTLMDLLKVE NELNGILCMK AITTLHKTFK ASLQEKVHPF IDIVIEIYSN IPQVVEEQFN G NQIDSKEN VDSTSRPNSP SFSSQSDDSK QLAQAMFSFK TLAESPITMV SLYSSYKELA ASSLGNFIPH VMKVLSLEVA KQ AEARKAA EEKGIILVNV CKEITNRANY GEFIIGQVKA ASFLAYLFIR RQAQTFLEPY QQAIPDIIIR LLQDCPSELS AAR KELLHA TRHILSTDFR KMFIPKIDLL FDLRVLIGEG FTAYETLRPL AYSTVADFIH NVRDHLTPAQ LWKSVSIYCK NLQD DSLAL TVQIMSAKLL LNLIEKIMRS ESKTESRQLL MVIIDAYTKR FKMLNSRYNG IMKQHATYEK EKQEKQNQER LLTNK LDGT TPSPSDDKKV ELIDEDQDVK MEDPTPEISD QETIKGDNDA STEPQDSEQQ LADFMSLQEY LPIQVSVPPE IDLLKD SRY LFKTLMTFLK TIMIGLKNSN PPSSQNHFNA QNWNETARGF SNEDINILKS LFRECILALR FFSTSKTSLP ASSMKQS FD ITGPNLPITS TKEEKDLMEI FATMFIHIDP ASFNEIVREE LPFMYKQMLD FASLLHIPQF FLASVITSSS FSGILITF L KSKLVDLGEV NIIKSNILIR LFKLCFMSVS LFPAANESVI LPHLNELILK SLKLSTTAKE PLVYFYLIRT LFRSIGGGR FENLYKEIMP LLQVLLESLS KLIHEARRPQ ERDIYVELCL TVPVRLSVLV PHLSYLMKPL VYALNGSQES VSQGLRTLEL CVDNLTAEY FDPIIEPVID DVMEALSKHL KPLPYYHQHS HTTLRILGKL GGRNRTFIKP VDNLKTDSEL FQNVEAMFKI H GLPNEVPL SITPGLSAAF SLLTDPRPRI HYRINSFKYI SGIFQLFLGA TQLPDDYANR LKESMDIILE DTIAPDEPLN KL HHFPVKD IAKYDSQMEL LVKLLESIFY AVSLQEVREE SKALIRGTCN HFILLYFNKM VIDKRKFVRK FSVDNHEGNL FLN ENCIFD AIIYALSSDN SAVRSMGLES VQLIYDSCVE LFGNIDCALK FAPLNVMCSK FIHCCFEEPY HKKLAGCIGL EMML NSLDI PMKYFNARQL EIIRALFYVL RDTAPELPCE VTNTAKRLIL NSLKEWNKEL TRNDVFSSVF QNLVSSLIVD LPNAN EIVR ATAQEALRTL SETTQVPIAT MISPCKHILL APIFGKPLRA LPFQMQIGNI DAITFCMGLE NSFLEYNEEL NRLVQE ALA LVDAEDESLV SAHRISEHKT SEQLVRLRVV CIQLLSLAIT KPEFAAAQQR SNIRVKILVV FFKSLCGRSI EIIRAAH GG LKAVIDLKMK LPKELLQNGL RPMLMNLSDH KKLTVASLEA LSGLLKLFIS YFKVGIGSKL LDHLLAWAQP RTLQQLGS Q DLENNSTVQI IVAILDVFHL LPPTAHKFMN DLMNALLYLE NNLHRCQYSP FREPLAKFLD RFPDESFEYF FNEFSKREI TTRFVYFVGL DSCSSLRAKV LESLPRVRGL LHQEGSAEEK CVRFSNLVDL CESLAASDKE WIKDKEELLG ELLDAGSVCL TLKRSSNVV SPLYFQVDQG FETLQLLYIE YFKSQPLGHE KVFNFIDKIS KEGLPFVLEF DDFIFNEVVK CQDIPTVQQT L DTIIRMTP QVSSLDARVY LYKRIFLPIC IYESEMHGDL SRLSQTENNE LPAWLKSFDS DVWKATGPLV DDYTSTLEDR YR LELMQLT ALLIKGAPTA LTDMRKDIIK FSWNYIKLDD NTSKQAAYVV TAYFISRFDT PSELTTRIFV ALLRCHQIDT RYL VKQALE LLAPVLSERT NSELDWLKWP RRVLSEDGFN ITQVANIYQL IVKFPDLFYP ARDHFIPNII TAMGKLTVMS NTSL ENQQL AIDLAELILK WETKLPKSEK LGSAEETEKE KSVSEDKMDI DVKEETKEDI AERPKAEDQI GGDDSDSSNI LTSED YEVS FAQREACVTF LIRYICISTQ RPSENELGKR ALNILYELLG PKYWSEVTVK LQFFERFLMS SDLNQPSLLG YCLNAL EVL AVALKWKPTT WIIENVSYLQ KLLEKCLRSD NQDIQEILQK VLGIILEAIN KETQGSEEDE PEEVTNFISL IVNIIGE DL SNMTSVAAGV SLCWTLSLYR PNALDSLLPS IMRTFNKLCR DHIAISLQGN QPQSGDFANI EFEAKVTTNL LEKILNLC A ARISSLDDQR RVFLSLLAQL IDRSVDKDML LKVINIVTEW IFKTDFYPTT KEKAGILGKM MIFDLRGEPE LSKKFNQVI VDIFESKELA HTELTARMET AFLFGTRLSD VSIRKKLMSI LSDSLELDID KRLFYIIKDQ NWEYLSDYPW LNQALQLLYG SFHLDSPIR LSPEENTLSP LQSITEGLAR EKSPVEKAPQ NIIDFVAKHN EFLDSVRSLT AGDILNPLID ISYQSAETIH N AWVVVFPV AYSAIESRYE LEFTRALVKL LFKDYHIRQQ DARPNVIKSL LDGVGKCPGL HLPPHLVKYL GSNYNAWYGA IK LLEELSE GQGIDNQKIS DANQDALLEV YMSLQEDDMF YGTWRRRAKY FETNAALSYE QIGIWDKALQ LYEAAQIKAR SGV FPFGES EYSLWEDHWI YCAEKLQHWE ILTELAKHEG FTDLLLECGW RGADWIADRE PLEQSVKTVM DIPTPRRQIF QTFL ALQGF SQQKDTLQDV SRLCDEGIQL TLRKWNALPQ RVTRAHIGLL HTFQQYVELM EASQVYSSLV TTNAQNLDVK SQELK RVLQ AWRERLPNVW DDINIWNDLV TWRQHVFGVI NRVYMPFVPV LQQSNGTNNG NSYAYRGYHE MAWVINRFAH VARKHE MPE VCINQLTKIY TLPNIEIQEA FLKLREQAKC HYQNSSELNT GLDVISNTNL VYFATQQKAE FFTLKGMFLA KLNAKDE AN QAFATAVQID LNLPKAWAEW GFFNDRRFKE NPEEIFHAKN AISCYLQAAG LYKDGKTRKL LCRILWLISL DDAAGSLA K TFEDHHGESP VWYWITFVPQ LLTSLSHKEA KIVRHILIQI AKSYPQSLHF QLRTTKEDYQ AIQRQAMAVN RAEEQSSNK QDTADSVLKN TNTPQPQTRT ETSGTTAESD KKPSIPPKEE QGSPQPSRPA TTQASPQAQS QENGESSQKH PPEIPTTDSR QPWQDVEEI MGILKTAYPL LALSLESLVD QLNQRFKCNA DEDAYRLVIV LYNDGVQQMN RVANPREEVK LPAATEASIS R FADSVLPK NIREVFEQDI IACNPNLETY ISKLRKWRDC LEEKLDRSYG KADLERVSLH LSLFHHQKFE DIEIPGQYLL HK DNNNHFI KIERFLPTLD LVRGSNGCYK RMTIRGNDGS LHPFAVQFPA ARHCRREERI FQLFRIFDDA LSRKVQSRRR NIS LTLPIA VPLSPHIRIL NDDKRYTTLM GIYEEFCRRK GQSRDEPFAY TIQKLRAAFD PRLPKPDIVS VRAEVLASIQ STLV PSTLL KDYYTEKFSN YENYWLFRKQ FTAQYASFIF MTYIMCINSR QPQKIHINEG SGNIWTSEML PTKVATGKTH STAYN NSTL DPAVKAGAPI FYNTESVPFR LTPNIQKFIG EAGLEGILSV YILVIANSLS DSEFDMEQYL SLFVRDEVIS WFAQQH RAS AQTNQLREIV RVNVELLTKR VLQLNHIPNS QNVATQFVLN LISQAVNPRN LAYTDSAWMA YL UniProtKB: SAGA and NuA4 histone acetyltransferase complexes subunits |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.018000000000000002 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blot for 1 second before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 80.0 K |
| Specialist optics | Spherical aberration corrector: Microscope has a Cs corrector |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Frames/image: 2-8 / Number grids imaged: 4 / Number real images: 8505 / Average exposure time: 1.0 sec. / Average electron dose: 60.0 e/Å2 Details: Images were collected in movie-mode at 17 frames per second, frame 1 was not acquired. Every two frames were joined together, producing 8 frames per second. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 3.4 µm / Calibrated defocus min: 1.4000000000000001 µm / Calibrated magnification: 127272 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.001 mm / Nominal defocus max: 3.4 µm / Nominal defocus min: 1.4000000000000001 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: B / Chain - Residue range: 1385-2549 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Secondary structure restraints were applied in Phenix. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-5oej: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)