+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2818 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Dengue virus serotype 2 with Fab fragments of human antibody B7 | |||||||||
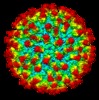
 Map data Map data | Reconstruction of Dengue Virus Serotype 2 in complex with Human Antibody B7 Fab Fragment. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | dengue virus / human monoclonal antibody / potent broad neutralization / E-dimer Dependent epitope | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Dengue virus 2 Dengue virus 2 | |||||||||
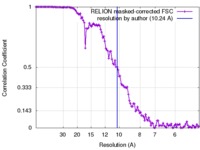
| Method | single particle reconstruction / cryo EM / Resolution: 10.24 Å | |||||||||
 Authors Authors | Dejnirattisai W / Wongwiwat W / Supasa S / Zhang X / Dai X / Rouvinsky A / Jumnainsong A / Edwards C / Quyen NT / Duangchinda T ...Dejnirattisai W / Wongwiwat W / Supasa S / Zhang X / Dai X / Rouvinsky A / Jumnainsong A / Edwards C / Quyen NT / Duangchinda T / Grimes JM / Tsai WY / Lai CY / Wang WK / Malasit P / Farrar J / Simmons CP / Zhou ZH / Rey FA / Mongkolsapaya J / Screaton GR | |||||||||
 Citation Citation |  Journal: Nat Immunol / Year: 2015 Journal: Nat Immunol / Year: 2015Title: A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Authors: Wanwisa Dejnirattisai / Wiyada Wongwiwat / Sunpetchuda Supasa / Xiaokang Zhang / Xinghong Dai / Alexander Rouvinski / Amonrat Jumnainsong / Carolyn Edwards / Nguyen Than Ha Quyen / Thaneeya ...Authors: Wanwisa Dejnirattisai / Wiyada Wongwiwat / Sunpetchuda Supasa / Xiaokang Zhang / Xinghong Dai / Alexander Rouvinski / Amonrat Jumnainsong / Carolyn Edwards / Nguyen Than Ha Quyen / Thaneeya Duangchinda / Jonathan M Grimes / Wen-Yang Tsai / Chih-Yun Lai / Wei-Kung Wang / Prida Malasit / Jeremy Farrar / Cameron P Simmons / Z Hong Zhou / Felix A Rey / Juthathip Mongkolsapaya / Gavin R Screaton /       Abstract: Dengue is a rapidly emerging, mosquito-borne viral infection, with an estimated 400 million infections occurring annually. To gain insight into dengue immunity, we characterized 145 human monoclonal ...Dengue is a rapidly emerging, mosquito-borne viral infection, with an estimated 400 million infections occurring annually. To gain insight into dengue immunity, we characterized 145 human monoclonal antibodies (mAbs) and identified a previously unknown epitope, the envelope dimer epitope (EDE), that bridges two envelope protein subunits that make up the 90 repeating dimers on the mature virion. The mAbs to EDE were broadly reactive across the dengue serocomplex and fully neutralized virus produced in either insect cells or primary human cells, with 50% neutralization in the low picomolar range. Our results provide a path to a subunit vaccine against dengue virus and have implications for the design and monitoring of future vaccine trials in which the induction of antibody to the EDE should be prioritized. #1:  Journal: NATURE Journal: NATURETitle: Recognition determinants of broadly neutralizing human antibodies against dengue viruses Authors: Rouvinski A / Guardado-Calvo P / Barba-Spaeth G / Duquerroy S / Vaney MC / Kikuti CM / Navarro Sanchez E / Dejnirattisai W / Wongwiwat W / Haouz A / Girard-Blanc C / Petres S / England P / ...Authors: Rouvinski A / Guardado-Calvo P / Barba-Spaeth G / Duquerroy S / Vaney MC / Kikuti CM / Navarro Sanchez E / Dejnirattisai W / Wongwiwat W / Haouz A / Girard-Blanc C / Petres S / England P / Supasa S / Shepard WE / Duangchinda T / Siridechadilok B / Despres P / Arenzana-Seisdedos F / Dussart P / Mongkolsapaya J / Screaton GR / Rey FA | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2818.map.gz emd_2818.map.gz | 59.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2818-v30.xml emd-2818-v30.xml emd-2818.xml emd-2818.xml | 13.2 KB 13.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_2818_fsc.xml emd_2818_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd-2818.png emd-2818.png | 364 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2818 http://ftp.pdbj.org/pub/emdb/structures/EMD-2818 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2818 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2818 | HTTPS FTP |
-Validation report
| Summary document |  emd_2818_validation.pdf.gz emd_2818_validation.pdf.gz | 318.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2818_full_validation.pdf.gz emd_2818_full_validation.pdf.gz | 317.2 KB | Display | |
| Data in XML |  emd_2818_validation.xml.gz emd_2818_validation.xml.gz | 12.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2818 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2818 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2818 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2818 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2818.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2818.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of Dengue Virus Serotype 2 in complex with Human Antibody B7 Fab Fragment. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.56 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Fab Fragment of human antibody to Dengue 2 virus
| Entire | Name: Fab Fragment of human antibody to Dengue 2 virus |
|---|---|
| Components |
|
-Supramolecule #1000: Fab Fragment of human antibody to Dengue 2 virus
| Supramolecule | Name: Fab Fragment of human antibody to Dengue 2 virus / type: sample / ID: 1000 / Number unique components: 2 |
|---|
-Supramolecule #1: Dengue virus 2
| Supramolecule | Name: Dengue virus 2 / type: virus / ID: 1 / NCBI-ID: 11060 / Sci species name: Dengue virus 2 / Sci species strain: NGC / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
| Virus shell | Shell ID: 1 / Diameter: 500 Å / T number (triangulation number): 3 |
-Macromolecule #1: FAB
| Macromolecule | Name: FAB / type: protein_or_peptide / ID: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: PBS |
|---|---|
| Grid | Details: 200 mesh quantifoil 2/2 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV / Method: Blot for 4 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Date | Jun 9, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Digitization - Sampling interval: 5.0 µm / Number real images: 1000 / Average electron dose: 25 e/Å2 Details: Every image is the average of 25 frames recorded by the direct electron detector Bits/pixel: 32 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.2 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B / Chain - #2 - Chain ID: C / Chain - #3 - Chain ID: D / Chain - #4 - Chain ID: E / Chain - #5 - Chain ID: F |
|---|---|
| Software | Name:  UCSF Chimera UCSF Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: cross coefficient |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)