+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23200 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
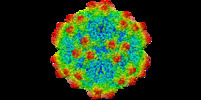
| Title | The genome-containing AAV12 capsid | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Icosahedral Capsid / AAV12 / Adeno-associated virus / Parvovirus / Gene Therapy / VIRUS | |||||||||
| Function / homology | Phospholipase A2-like domain / Phospholipase A2-like domain / Parvovirus coat protein VP2 / Parvovirus coat protein VP1/VP2 / Parvovirus coat protein VP2 / Capsid/spike protein, ssDNA virus / T=1 icosahedral viral capsid / structural molecule activity / VP1 Function and homology information Function and homology information | |||||||||
| Biological species |  Adeno-associated virus 12 Adeno-associated virus 12 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.67 Å | |||||||||
 Authors Authors | Mietzsch M / Agbandje-McKenna M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Viruses / Year: 2021 Journal: Viruses / Year: 2021Title: Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features. Authors: Mario Mietzsch / Ariana Jose / Paul Chipman / Nilakshee Bhattacharya / Nadia Daneshparvar / Robert McKenna / Mavis Agbandje-McKenna /  Abstract: The capsid structures of most Adeno-associated virus (AAV) serotypes, already assigned to an antigenic clade, have been previously determined. This study reports the remaining capsid structures of ...The capsid structures of most Adeno-associated virus (AAV) serotypes, already assigned to an antigenic clade, have been previously determined. This study reports the remaining capsid structures of AAV7, AAV11, AAV12, and AAV13 determined by cryo-electron microscopy and three-dimensional image reconstruction to 2.96, 2.86, 2.54, and 2.76 Å resolution, respectively. These structures complete the structural atlas of the AAV serotype capsids. AAV7 represents the first clade D capsid structure; AAV11 and AAV12 are of a currently unassigned clade that would include AAV4; and AAV13 represents the first AAV2-AAV3 hybrid clade C capsid structure. These newly determined capsid structures all exhibit the AAV capsid features including 5-fold channels, 3-fold protrusions, 2-fold depressions, and a nucleotide binding pocket with an ordered nucleotide in genome-containing capsids. However, these structures have viral proteins that display clade-specific loop conformations. This structural characterization completes our three-dimensional library of the current AAV serotypes to provide an atlas of surface loop configurations compatible with capsid assembly and amenable for future vector engineering efforts. Derived vectors could improve gene delivery success with respect to specific tissue targeting, transduction efficiency, antigenicity or receptor retargeting. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23200.map.gz emd_23200.map.gz | 262.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23200-v30.xml emd-23200-v30.xml emd-23200.xml emd-23200.xml | 9.8 KB 9.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23200.png emd_23200.png | 323.8 KB | ||
| Filedesc metadata |  emd-23200.cif.gz emd-23200.cif.gz | 5.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23200 http://ftp.pdbj.org/pub/emdb/structures/EMD-23200 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23200 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23200 | HTTPS FTP |
-Validation report
| Summary document |  emd_23200_validation.pdf.gz emd_23200_validation.pdf.gz | 703.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23200_full_validation.pdf.gz emd_23200_full_validation.pdf.gz | 703 KB | Display | |
| Data in XML |  emd_23200_validation.xml.gz emd_23200_validation.xml.gz | 7.6 KB | Display | |
| Data in CIF |  emd_23200_validation.cif.gz emd_23200_validation.cif.gz | 8.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23200 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23200 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23200 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23200 | HTTPS FTP |
-Related structure data
| Related structure data |  7l6aMC  7l5qC  7l5uC  7l6bC  7l6eC  7l6fC  7l6hC  7l6iC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23200.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23200.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.082 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Adeno-associated virus 12
| Entire | Name:  Adeno-associated virus 12 Adeno-associated virus 12 |
|---|---|
| Components |
|
-Supramolecule #1: Adeno-associated virus 12
| Supramolecule | Name: Adeno-associated virus 12 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 235458 / Sci species name: Adeno-associated virus 12 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: VP1
| Macromolecule | Name: VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 60 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Adeno-associated virus 12 Adeno-associated virus 12 |
| Molecular weight | Theoretical: 58.549543 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DGVGNASGDW HCDSTWSEGR VTTTSTRTWV LPTYNNHLYL RIGTTANSNT YNGFSTPWGY FDFNRFHCHF SPRDWQRLIN NNWGLRPKS MRVKIFNIQV KEVTTSNGET TVANNLTSTV QIFADSTYEL PYVMDAGQEG SFPPFPNDVF MVPQYGYCGV V TGKNQNQT ...String: DGVGNASGDW HCDSTWSEGR VTTTSTRTWV LPTYNNHLYL RIGTTANSNT YNGFSTPWGY FDFNRFHCHF SPRDWQRLIN NNWGLRPKS MRVKIFNIQV KEVTTSNGET TVANNLTSTV QIFADSTYEL PYVMDAGQEG SFPPFPNDVF MVPQYGYCGV V TGKNQNQT DRNAFYCLEY FPSQMLRTGN NFEVSYQFEK VPFHSMYAHS QSLDRMMNPL LDQYLWHLQS TTTGNSLNQG TA TTTYGKI TTGDFAYYRK NWLPGACIKQ QKFSKNANQN YKIPASGGDA LLKYDTHTTL NGRWSNMAPG PPMATAGAGD SDF SNSQLI FAGPNPSGNT TTSSNNLLFT SEEEIATTNP RDTDMFGQIA DNNQNATTAP HIANLDAMGI VPGMVWQNRD IYYQ GPIWA KVPHTDGHFH PSPLMGGFGL KHPPPQIFIK NTPVPANPNT TFSAARINSF LTQYSTGQVA VQIDWEIQKE HSKRW NPEV QFTSNYGTQN SMLWAPDNAG NYHELRAIGS RFLTHHL UniProtKB: VP1 |
-Macromolecule #2: 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE
| Macromolecule | Name: 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE / type: ligand / ID: 2 / Number of copies: 60 / Formula: D5M |
|---|---|
| Molecular weight | Theoretical: 331.222 Da |
| Chemical component information | 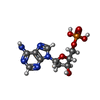 ChemComp-D5M: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.67 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cisTEM / Number images used: 40764 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: RANDOM ASSIGNMENT |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)





















