[English] 日本語
 Yorodumi
Yorodumi- EMDB-22030: Reprocessing of EMPIAR-10335 data (streptavidin) with particles p... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22030 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Reprocessing of EMPIAR-10335 data (streptavidin) with particles picked by Kpicker | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Streptomyces avidinii (bacteria) Streptomyces avidinii (bacteria) | |||||||||
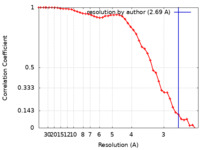
| Method | single particle reconstruction / cryo EM / Resolution: 2.69 Å | |||||||||
 Authors Authors | McSweeney DM / McSweeney SM / Liu Q | |||||||||
 Citation Citation |  Journal: IUCrJ / Year: 2020 Journal: IUCrJ / Year: 2020Title: A self-supervised workflow for particle picking in cryo-EM. Authors: Donal M McSweeney / Sean M McSweeney / Qun Liu /  Abstract: High-resolution single-particle cryo-EM data analysis relies on accurate particle picking. To facilitate the particle picking process, a self-supervised workflow has been developed. This includes an ...High-resolution single-particle cryo-EM data analysis relies on accurate particle picking. To facilitate the particle picking process, a self-supervised workflow has been developed. This includes an iterative strategy, which uses a 2D class average to improve training particles, and a progressively improved convolutional neural network for particle picking. To automate the selection of particles, a threshold is defined (%/Res) using the ratio of percentage class distribution and resolution as a cutoff. This workflow has been tested using six publicly available data sets with different particle sizes and shapes, and can automatically pick particles with minimal user input. The picked particles support high-resolution reconstructions at 3.0 Å or better. This workflow is a step towards automated single-particle cryo-EM data analysis at the stage of particle picking. It may be used in conjunction with commonly used single-particle analysis packages such as , , , and . | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22030.map.gz emd_22030.map.gz | 1.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22030-v30.xml emd-22030-v30.xml emd-22030.xml emd-22030.xml | 8.1 KB 8.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22030_fsc.xml emd_22030_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_22030.png emd_22030.png | 189.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22030 http://ftp.pdbj.org/pub/emdb/structures/EMD-22030 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22030 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22030 | HTTPS FTP |
-Validation report
| Summary document |  emd_22030_validation.pdf.gz emd_22030_validation.pdf.gz | 78.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22030_full_validation.pdf.gz emd_22030_full_validation.pdf.gz | 77.5 KB | Display | |
| Data in XML |  emd_22030_validation.xml.gz emd_22030_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22030 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22030 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22030 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22030 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22030.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22030.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.072 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Streptavidin
| Entire | Name: Streptavidin |
|---|---|
| Components |
|
-Supramolecule #1: Streptavidin
| Supramolecule | Name: Streptavidin / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Streptomyces avidinii (bacteria) Streptomyces avidinii (bacteria) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 49.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)