+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | TRRAP in human Tip60 complex locally refined 1 | |||||||||
 Map data Map data | TRRAPP locally refined 1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Eukaryotic transcription / Histone acetyltransferase / chromatin remodeling / Complex / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationtranscription factor TFTC complex / Swr1 complex / protein antigen binding / regulation of double-strand break repair / SAGA complex / ATP-dependent chromatin remodeler activity / Formation of Senescence-Associated Heterochromatin Foci (SAHF) / NuA4 histone acetyltransferase complex / regulation of RNA splicing / positive regulation of double-strand break repair via homologous recombination ...transcription factor TFTC complex / Swr1 complex / protein antigen binding / regulation of double-strand break repair / SAGA complex / ATP-dependent chromatin remodeler activity / Formation of Senescence-Associated Heterochromatin Foci (SAHF) / NuA4 histone acetyltransferase complex / regulation of RNA splicing / positive regulation of double-strand break repair via homologous recombination / regulation of DNA repair / helicase activity / transcription coregulator activity / Formation of the beta-catenin:TCF transactivating complex / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / DNA Damage/Telomere Stress Induced Senescence / nucleosome / chromatin organization / HATs acetylate histones / regulation of apoptotic process / regulation of cell cycle / Ub-specific processing proteases / hydrolase activity / nuclear speck / DNA repair / chromatin binding / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / positive regulation of DNA-templated transcription / Golgi apparatus / DNA binding / nucleoplasm / ATP binding / nucleus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
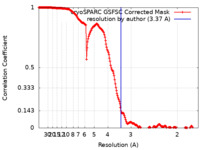
| Method | single particle reconstruction / cryo EM / Resolution: 3.37 Å | |||||||||
 Authors Authors | Li C / Smirnova E / Schnitzler C / Crucifix C / Concordet JP / Brion A / Poterszman A / Schultz P / Papai G / Ben-Shem A | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structure of the human TIP60-C histone exchange and acetyltransferase complex. Authors: Changqing Li / Ekaterina Smirnova / Charlotte Schnitzler / Corinne Crucifix / Jean Paul Concordet / Alice Brion / Arnaud Poterszman / Patrick Schultz / Gabor Papai / Adam Ben-Shem /  Abstract: Chromatin structure is a key regulator of DNA transcription, replication and repair. In humans, the TIP60-EP400 complex (TIP60-C) is a 20-subunit assembly that affects chromatin structure through two ...Chromatin structure is a key regulator of DNA transcription, replication and repair. In humans, the TIP60-EP400 complex (TIP60-C) is a 20-subunit assembly that affects chromatin structure through two enzymatic activities: ATP-dependent exchange of histone H2A-H2B for H2A.Z-H2B, and histone acetylation. In yeast, however, these activities are performed by two independent complexes-SWR1 and NuA4, respectively. How the activities of the two complexes are merged into one supercomplex in humans, and what this association entails for the structure and mechanism of the proteins and their recruitment to chromatin, are unknown. Here we describe the structure of the endogenous human TIP60-C. We find a three-lobed architecture composed of SWR1-like (SWR1L) and NuA4-like (NuA4L) parts, which associate with a TRRAP activator-binding module. The huge EP400 subunit contains the ATPase motor, traverses the junction between SWR1L and NuA4L twice and constitutes the scaffold of the three-lobed architecture. NuA4L is completely rearranged compared with its yeast counterpart. TRRAP is flexibly tethered to NuA4L-in stark contrast to its robust connection to the completely opposite side of NuA4 in yeast. A modelled nucleosome bound to SWR1L, supported by tests of TIP60-C activity, suggests that some aspects of the histone exchange mechanism diverge from what is seen in yeast. Furthermore, a fixed actin module (as opposed to the mobile actin subcomplex in SWR1; ref. ), the flexibility of TRRAP and the weak effect of extranucleosomal DNA on exchange activity lead to a different, activator-based mode of enlisting TIP60-C to chromatin. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18613.map.gz emd_18613.map.gz | 334.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18613-v30.xml emd-18613-v30.xml emd-18613.xml emd-18613.xml | 23.2 KB 23.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18613_fsc.xml emd_18613_fsc.xml | 18.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_18613.png emd_18613.png | 63.5 KB | ||
| Masks |  emd_18613_msk_1.map emd_18613_msk_1.map | 669.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18613.cif.gz emd-18613.cif.gz | 9 KB | ||
| Others |  emd_18613_half_map_1.map.gz emd_18613_half_map_1.map.gz emd_18613_half_map_2.map.gz emd_18613_half_map_2.map.gz | 621.6 MB 621.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18613 http://ftp.pdbj.org/pub/emdb/structures/EMD-18613 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18613 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18613 | HTTPS FTP |
-Validation report
| Summary document |  emd_18613_validation.pdf.gz emd_18613_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18613_full_validation.pdf.gz emd_18613_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_18613_validation.xml.gz emd_18613_validation.xml.gz | 28 KB | Display | |
| Data in CIF |  emd_18613_validation.cif.gz emd_18613_validation.cif.gz | 36.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18613 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18613 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18613 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18613 | HTTPS FTP |
-Related structure data
| Related structure data |  8qr1C  8qriC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18613.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18613.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TRRAPP locally refined 1 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.862 Å | ||||||||||||||||||||||||||||||||||||
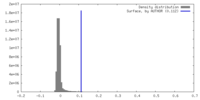
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18613_msk_1.map emd_18613_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
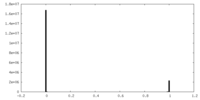
| Density Histograms |
-Half map: Half map A
| File | emd_18613_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
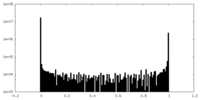
| Density Histograms |
-Half map: Half map B
| File | emd_18613_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
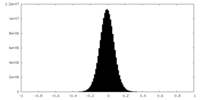
| Density Histograms |
- Sample components
Sample components
-Entire : Human Tip60 complex
| Entire | Name: Human Tip60 complex |
|---|---|
| Components |
|
-Supramolecule #1: Human Tip60 complex
| Supramolecule | Name: Human Tip60 complex / type: cell / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: TRRAP
| Macromolecule | Name: TRRAP / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAFVATQGAT VVDQTTLMKK YLQFVAALTD VNTPDETKLK MMQEVSENFE NVTSSPQYST FLEHIIPRFL TFLQDGEVQF LQEKPAQQLR KLVLEIIHRI PTNEHLRPHT KNVLSVMFRF LETENEENVL ICLRIIIELH KQFRPPITQE IHHFLDFVKQ IYKELPKVVN ...String: MAFVATQGAT VVDQTTLMKK YLQFVAALTD VNTPDETKLK MMQEVSENFE NVTSSPQYST FLEHIIPRFL TFLQDGEVQF LQEKPAQQLR KLVLEIIHRI PTNEHLRPHT KNVLSVMFRF LETENEENVL ICLRIIIELH KQFRPPITQE IHHFLDFVKQ IYKELPKVVN RYFENPQVIP ENTVPPPEMV GMITTIAVKV NPEREDSETR THSIIPRGSL SLKVLAELPI IVVLMYQLYK LNIHNVVAEF VPLIMNTIAI QVSAQARQHK LYNKELYADF IAAQIKTLSF LAYIIRIYQE LVTKYSQQMV KGMLQLLSNC PAETAHLRKE LLIAAKHILT TELRNQFIPC MDKLFDESIL IGSGYTARET LRPLAYSTLA DLVHHVRQHL PLSDLSLAVQ LFAKNIDDES LPSSIQTMSC KLLLNLVDCI RSKSEQESGN GRDVLMRMLE VFVLKFHTIA RYQLSAIFKK CKPQSELGAV EAALPGVPTA PAAPGPAPSP APVPAPPPPP PPPPPATPVT PAPVPPFEKQ GEKDKEDKQT FQVTDCRSLV KTLVCGVKTI TWGITSCKAP GEAQFIPNKQ LQPKETQIYI KLVKYAMQAL DIYQVQIAGN GQTYIRVANC QTVRMKEEKE VLEHFAGVFT MMNPLTFKEI FQTTVPYMVE RISKNYALQI VANSFLANPT TSALFATILV EYLLDRLPEM GSNVELSNLY LKLFKLVFGS VSLFAAENEQ MLKPHLHKIV NSSMELAQTA KEPYNYFLLL RALFRSIGGG SHDLLYQEFL PLLPNLLQGL NMLQSGLHKQ HMKDLFVELC LTVPVRLSSL LPYLPMLMDP LVSALNGSQT LVSQGLRTLE LCVDNLQPDF LYDHIQPVRA ELMQALWRTL RNPADSISHV AYRVLGKFGG SNRKMLKESQ KLHYVVTEVQ GPSITVEFSD CKASLQLPME KAIETALDCL KSANTEPYYR RQAWEVIKCF LVAMMSLEDN KHALYQLLAH PNFTEKTIPN VIISHRYKAQ DTPARKTFEQ ALTGAFMSAV IKDLRPSALP FVASLIRHYT MVAVAQQCGP FLLPCYQVGS QPSTAMFHSE ENGSKGMDPL VLIDAIAICM AYEEKELCKI GEVALAVIFD VASIILGSKE RACQLPLFSY IVERLCACCY EQAWYAKLGG VVSIKFLMER LPLTWVLQNQ QTFLKALLFV MMDLTGEVSN GAVAMAKTTL EQLLMRCATP LKDEERAEEI VAAQEKSFHH VTHDLVREVT SPNSTVRKQA MHSLQVLAQV TGKSVTVIME PHKEVLQDMV PPKKHLLRHQ PANAQIGLME GNTFCTTLQP RLFTMDLNVV EHKVFYTELL NLCEAEDSAL TKLPCYKSLP SLVPLRIAAL NALAACNYLP QSREKIIAAL FKALNSTNSE LQEAGEACMR KFLEGATIEV DQIHTHMRPL LMMLGDYRSL TLNVVNRLTS VTRLFPNSFN DKFCDQMMQH LRKWMEVVVI THKGGQRSDG NESISECGRC PLSPFCQFEE MKICSAIINL FHLIPAAPQT LVKPLLEVVM KTERAMLIEA GSPFREPLIK FLTRHPSQTV ELFMMEATLN DPQWSRMFMS FLKHKDARPL RDVLAANPNR FITLLLPGGA QTAVRPGSPS TSTMRLDLQF QAIKIISIIV KNDDSWLASQ HSLVSQLRRV WVSENFQERH RKENMAATNW KEPKLLAYCL LNYCKRNYGD IELLFQLLRA FTGRFLCNMT FLKEYMEEEI PKNYSIAQKR ALFFRFVDFN DPNFGDELKA KVLQHILNPA FLYSFEKGEG EQLLGPPNPE GDNPESITSV FITKVLDPEK QADMLDSLRI YLLQYATLLV EHAPHHIHDN NKNRNSKLRR LMTFAWPCLL SKACVDPACK YSGHLLLAHI IAKFAIHKKI VLQVFHSLLK AHAMEARAIV RQAMAILTPA VPARMEDGHQ MLTHWTRKII VEEGHTVPQL VHILHLIVQH FKVYYPVRHH LVQHMVSAMQ RLGFTPSVTI EQRRLAVDLS EVVIKWELQR IKDQQPDSDM DPNSSGEGVN SVSSSIKRGL SVDSAQEVKR FRTATGAISA VFGRSQSLPG ADSLLAKPID KQHTDTVVNF LIRVACQVND NTNTAGSPGE VLSRRCVNLL KTALRPDMWP KSELKLQWFD KLLMTVEQPN QVNYGNICTG LEVLSFLLTV LQSPAILSSF KPLQRGIAAC MTCGNTKVLR AVHSLLSRLM SIFPTEPSTS SVASKYEELE CLYAAVGKVI YEGLTNYEKA TNANPSQLFG TLMILKSACS NNPSYIDRLI SVFMRSLQKM VREHLNPQAA SGSTEATSGT SELVMLSLEL VKTRLAVMSM EMRKNFIQAI LTSLIEKSPD AKILRAVVKI VEEWVKNNSP MAANQTPTLR EKSILLVKMM TYIEKRFPED LELNAQFLDL VNYVYRDETL SGSELTAKLE PAFLSGLRCA QPLIRAKFFE VFDNSMKRRV YERLLYVTCS QNWEAMGNHF WIKQCIELLL AVCEKSTPIG TSCQGAMLPS ITNVINLADS HDRAAFAMVT HVKQEPRERE NSESKEEDVE IDIELAPGDQ TSTPKTKELS EKDIGNQLHM LTNRHDKFLD TLREVKTGAL LSAFVQLCHI STTLAEKTWV QLFPRLWKIL SDRQQHALAG EISPFLCSGS HQVQRDCQPS ALNCFVEAMS QCVPPIPIRP CVLKYLGKTH NLWFRSTLML EHQAFEKGLS LQIKPKQTTE FYEQESITPP QQEILDSLAE LYSLLQEEDM WAGLWQKRCK YSETATAIAY EQHGFFEQAQ ESYEKAMDKA KKEHERSNAS PAIFPEYQLW EDHWIRCSKE LNQWEALTEY GQSKGHINPY LVLECAWRVS NWTAMKEALV QVEVSCPKEM AWKVNMYRGY LAICHPEEQQ LSFIERLVEM ASSLAIREWR RLPHVVSHVH TPLLQAAQQI IELQEAAQIN AGLQPTNLGR NNSLHDMKTV VKTWRNRLPI VSDDLSHWSS IFMWRQHHYQ GKPTWSGMHS SSIVTAYENS SQHDPSSNNA MLGVHASASA IIQYGKIARK QGLVNVALDI LSRIHTIPTV PIVDCFQKIR QQVKCYLQLA GVMGKNECMQ GLEVIESTNL KYFTKEMTAE FYALKGMFLA QINKSEEANK AFSAAVQMHD VLVKAWAMWG DYLENIFVKE RQLHLGVSAI TCYLHACRHQ NESKSRKYLA KVLWLLSFDD DKNTLADAVD KYCIGVPPIQ WLAWIPQLLT CLVGSEGKLL LNLISQVGRV YPQAVYFPIR TLYLTLKIEQ RERYKSDPGP IRATAPMWRC SRIMHMQREL HPTLLSSLEG IVDQMVWFRE NWHEEVLRQL QQGLAKCYSV AFEKSGAVSD AKITPHTLNF VKKLVSTFGV GLENVSNVST MFSSAASESL ARRAQATAQD PVFQKLKGQF TTDFDFSVPG SMKLHNLISK LKKWIKILEA KTKQLPKFFL IEEKCRFLSN FSAQTAEVEI PGEFLMPKPT HYYIKIARFM PRVEIVQKHN TAARRLYIRG HNGKIYPYLV MNDACLTESR REERVLQLLR LLNPCLEKRK ETTKRHLFFT VPRVVAVSPQ MRLVEDNPSS LSLVEIYKQR CAKKGIEHDN PISRYYDRLA TVQARGTQAS HQVLRDILKE VQSNMVPRSM LKEWALHTFP NATDYWTFRK MFTIQLALIG FAEFVLHLNR LNPEMLQIAQ DTGKLNVAYF RFDINDATGD LDANRPVPFR LTPNISEFLT TIGVSGPLTA SMIAVARCFA QPNFKVDGIL KTVLRDEIIA WHKKTQEDTS SPLSAAGQPE NMDSQQLVSL VQKAVTAIMT RLHNLAQFEG GESKVNTLVA AANSLDNLCR MDPAWHPWL UniProtKB: Transformation/transcription domain-associated protein |
-Macromolecule #2: EP400
| Macromolecule | Name: EP400 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MHHGTGPQNV QHQLQRSRAC PGSEGEEQPA HPNPPPSPAA PFAPSASPSA PQSPSYQIQQ LMNRSPATGQ NVNITLQSVG PVVGGNQQIT LAPLPLPSPT SPGFQFSAQP RRFEHGSPSY IQVTSPLSQQ VQTQSPTQPS PGPGQALQNV RAGAPGPGLG LCSSSPTGGF ...String: MHHGTGPQNV QHQLQRSRAC PGSEGEEQPA HPNPPPSPAA PFAPSASPSA PQSPSYQIQQ LMNRSPATGQ NVNITLQSVG PVVGGNQQIT LAPLPLPSPT SPGFQFSAQP RRFEHGSPSY IQVTSPLSQQ VQTQSPTQPS PGPGQALQNV RAGAPGPGLG LCSSSPTGGF VDASVLVRQI SLSPSSGGHF VFQDGSGLTQ IAQGAQVQLQ HPGTPITVRE RRPSQPHTQS GGTIHHLGPQ SPAAAGGAGL QPLASPSHIT TANLPPQISS IIQGQLVQQQ QVLQGPPLPR PLGFERTPGV LLPGAGGAAG FGMTSPPPPT SPSRTAVPPG LSSLPLTSVG NTGMKKVPKK LEEIPPASPE MAQMRKQCLD YHYQEMQALK EVFKEYLIEL FFLQHFQGNM MDFLAFKKKH YAPLQAYLRQ NDLDIEEEEE EEEEEEEKSE VINDEVKVVT GKDGQTGTPV AIATQLPPKV SAAFSSQQQP FQQALAGSLV AGAGSTVETD LFKRQQAMPS TGMAEQSKRP RLEVGHQGVV FQHPGADAGV PLQQLMPTAQ GGMPPTPQAA QLAGQRQSQQ QYDPSTGPPV QNAASLHTPL PQLPGRLPPA GVPTAALSSA LQFAQQPQVV EAQTQLQIPV KTQQPNVPIP APPSSQLPIP PSQPAQLALH VPTPGKVQVQ ASQLSSLPQM VASTRLPVDP APPCPRPLPT SSTSSLAPVS GSGPGPSPAR SSPVNRPSSA TNKALSPVTS RTPGVVASAP TKPQSPAQNA TSSQDSSQDT LTEQITLENQ VHQRIAELRK AGLWSQRRLP KLQEAPRPKS HWDYLLEEMQ WMATDFAQER RWKVAAAKKL VRTVVRHHEE KQLREERGKK EEQSRLRRIA ASTAREIECF WSNIEQVVEI KLRVELEEKR KKALNLQKVS RRGKELRPKG FDALQESSLD SGMSGRKRKA SISLTDDEVD DEEETIEEEE ANEGVVDHQT ELSNLAKEAE LPLLDLMKLY EGAFLPSSQW PRPKPDGEDT SGEEDADDCP GDRESRKDLV LIDSLFIMDQ FKAAERMNIG KPNAKDIADV TAVAEAILPK GSARVTTSVK FNAPSLLYGA LRDYQKIGLD WLAKLYRKNL NGILADEAGL GKTVQIIAFF AHLACNEGNW GPHLVVVRSC NILKWELELK RWCPGLKILS YIGSHRELKA KRQEWAEPNS FHVCITSYTQ FFRGLTAFTR VRWKCLVIDE MQRVKGMTER HWEAVFTLQS QQRLLLIDSP LHNTFLELWT MVHFLVPGIS RPYLSSPLRA PSEESQDYYH KVVIRLHRVT QPFILRRTKR DVEKQLTKKY EHVLKCRLSN RQKALYEDVI LQPGTQEALK SGHFVNVLSI LVRLQRICNH PGLVEPRHPG SSYVAGPLEY PSASLILKAL ERDFWKEADL SMFDLIGLEN KITRHEAELL SKKKIPRKLM EEISTSAAPA ARPAAAKLKA SRLFQPVQYG QKPEGRTVAF PSTHPPRTAA PTTASAAPQG PLRGRPPIAT FSANPEAKAA AAPFQTSQAS ASAPRHQPAS ASSTAASPAH PAKLRAQTTA QASTPGQPPP QPQAPSHAAG QSALPQRLVL PSQAQARLPS GEVVKIAQLA SITGPQSRVA QPETPVTLQF QGSKFTLSHS QLRQLTAGQP LQLQGSVLQI VSAPGQPYLR APGPVVMQTV SQAGAVHGAL GSKPPAGGPS PAPLTPQVGV PGRVAVNALA VGEPGTASKP ASPIGGPTQE EKTRLLKERL DQIYLVNERR CSQAPVYGRD LLRICALPSH GRVQWRGSLD GRRGKEAGPA HSYTSSSESP SELMLTLCRC GESLQDVIDR VAFVIPPVVA APPSLRVPRP PPLYSHRMRI LRQGLREHAA PYFQQLRQTT APRLLQFPEL RLVQFDSGKL EALAILLQKL KSEGRRVLIL SQMILMLDIL EMFLNFHYLT YVRIDENASS EQRQELMRSF NRDRRIFCAI LSTHSRTTGI NLVEADTVVF YDNDLNPVMD AKAQEWCDRI GRCKDIHIYR LVSGNSIEEK LLKNGTKDLI REVAAQGNDY SMAFLTQRTI QELFEVYSPM DDAGFPVKAE EFVVLSQEPS VTETIAPKIA RPFIEALKSI EYLEEDAQKS AQEGVLGPHT DALSSDSENM PCDEEPSQLE ELADFMEQLT PIEKYALNYL ELFHTSIEQE KERNSEDAVM TAVRAWEFWN LKTLQEREAR LRLEQEEAEL LTYTREDAYS MEYVYEDVDG QTEVMPLWTP PTPPQDDSDI YLDSVMCLMY EATPIPEAKL PPVYVRKERK RHKTDPSAAG RKKKQRHGEA VVPPRSLFDR ATPGLLKIRR EGKEQKKNIL LKQQVPFAKP LPTFAKPTAE PGQDNPEWLI SEDWALLQAV KQLLELPLNL TIVSPAHTPN WDLVSDVVNS CSRIYRSSKQ CRNRYENVII PREEGKSKNN RPLRTSQIYA QDENATHTQL YTSHFDLMKM TAGKRSPPIK PLLGMNPFQK NPKHASVLAE SGINYDKPLP PIQVASLRAE RIAKEKKALA DQQKAQQPAV AQPPPPQPQP PPPPQQPPPP LPQPQAAGSQ PPAGPPAVQP QPQPQPQTQP QPVQAPAKAQ PAITTGGSAA VLAGTIKTSV TGTSMPTGAV SGNVIVNTIA GVPAATFQSI NKRLASPVAP GALTTPGGSA PAQVVHTQPP PRAVGSPATA TPDLVSMATT QGVRAVTSVT ASAVVTTNLT PVQTPARSLV PQVSQATGVQ LPGKTITPAH FQLLRQQQQQ QQQQQQQQQQ QQQQQQQQQQ QQQQTTTTSQ VQVPQIQGQA QSPAQIKAVG KLTPEHLIKM QKQKLQMPPQ PPPPQAQSAP PQPTAQVQVQ TSQPPQQQSP QLTTVTAPRP GALLTGTTVA NLQVARLTRV PTSQLQAQGQ MQTQAPQPAQ VALAKPPVVS VPAAVVSSPG VTTLPMNVAG ISVAIGQPQK AAGQTVVAQP VHMQQLLKLK QQAVQQQKAI QPQAAQGPAA VQQKITAQQI TTPGAQQKVA YAAQPALKTQ FLTTPISQAQ KLAGAQQVQT QIQVAKLPQV VQQQTPVASI QQVASASQQA SPQTVALTQA TAAGQQVQMI PAVTATAQVV QQKLIQQQVV TTASAPLQTP GAPNPAQVPA SSDSPSQQPK LQMRVPAVRL KTPTKPPCQ UniProtKB: E1A-binding protein p400 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 279 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)