+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human Tip60 complex | |||||||||
 Map data Map data | Composite map of the human Tip60 complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Eukaryotic transcription / Histone acetyltransferase / chromatin remodeling / Complex / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationpiccolo histone acetyltransferase complex / promoter-enhancer loop anchoring activity / telomerase RNA localization to Cajal body / regulation of DNA strand elongation / positive regulation of telomere maintenance in response to DNA damage / positive regulation of norepinephrine uptake / histone chaperone activity / sperm DNA condensation / establishment of protein localization to chromatin / R2TP complex ...piccolo histone acetyltransferase complex / promoter-enhancer loop anchoring activity / telomerase RNA localization to Cajal body / regulation of DNA strand elongation / positive regulation of telomere maintenance in response to DNA damage / positive regulation of norepinephrine uptake / histone chaperone activity / sperm DNA condensation / establishment of protein localization to chromatin / R2TP complex / dynein axonemal particle / bBAF complex / cellular response to cytochalasin B / neural retina development / npBAF complex / nBAF complex / brahma complex / regulation of transepithelial transport / morphogenesis of a polarized epithelium / structural constituent of postsynaptic actin cytoskeleton / Formation of annular gap junctions / Formation of the dystrophin-glycoprotein complex (DGC) / RPAP3/R2TP/prefoldin-like complex / GBAF complex / Swr1 complex / Gap junction degradation / Folding of actin by CCT/TriC / regulation of G0 to G1 transition / protein localization to adherens junction / Cell-extracellular matrix interactions / protein antigen binding / dense body / Tat protein binding / postsynaptic actin cytoskeleton / Ino80 complex / Prefoldin mediated transfer of substrate to CCT/TriC / RSC-type complex / blastocyst formation / chromatin-protein adaptor activity / regulation of double-strand break repair / regulation of nucleotide-excision repair / Adherens junctions interactions / RHOF GTPase cycle / adherens junction assembly / apical protein localization / box C/D snoRNP assembly / Sensory processing of sound by inner hair cells of the cochlea / Sensory processing of sound by outer hair cells of the cochlea / protein folding chaperone complex / Interaction between L1 and Ankyrins / tight junction / SWI/SNF complex / regulation of mitotic metaphase/anaphase transition / positive regulation of T cell differentiation / Formation of Senescence-Associated Heterochromatin Foci (SAHF) / apical junction complex / positive regulation of double-strand break repair / spinal cord development / negative regulation of gene expression, epigenetic / maintenance of blood-brain barrier / regulation of norepinephrine uptake / regulation of chromosome organization / nitric-oxide synthase binding / transporter regulator activity / cortical cytoskeleton / Transcriptional Regulation by E2F6 / establishment or maintenance of cell polarity / positive regulation of stem cell population maintenance / NuA4 histone acetyltransferase complex / Recycling pathway of L1 / Regulation of MITF-M-dependent genes involved in pigmentation / RUNX1 interacts with co-factors whose precise effect on RUNX1 targets is not known / TFIID-class transcription factor complex binding / regulation of DNA replication / brush border / MLL1 complex / regulation of G1/S transition of mitotic cell cycle / regulation of embryonic development / somatic stem cell population maintenance / EPH-ephrin mediated repulsion of cells / negative regulation of cell differentiation / Telomere Extension By Telomerase / kinesin binding / spermatid development / RHO GTPases Activate WASPs and WAVEs / regulation of synaptic vesicle endocytosis / enzyme-substrate adaptor activity / positive regulation of myoblast differentiation / RHO GTPases activate IQGAPs / RNA polymerase II core promoter sequence-specific DNA binding / regulation of DNA repair / regulation of protein localization to plasma membrane / positive regulation of double-strand break repair via homologous recombination / EPHB-mediated forward signaling / cytoskeleton organization / Deposition of new CENPA-containing nucleosomes at the centromere / substantia nigra development / transcription initiation-coupled chromatin remodeling / axonogenesis / DNA helicase activity Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.4 Å | |||||||||
 Authors Authors | Li C / Smirnova E / Schnitzler C / Crucifix C / Concordet JP / Brion A / Poterszman A / Schultz P / Papai G / Ben-Shem A | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structure of the human TIP60-C histone exchange and acetyltransferase complex. Authors: Changqing Li / Ekaterina Smirnova / Charlotte Schnitzler / Corinne Crucifix / Jean Paul Concordet / Alice Brion / Arnaud Poterszman / Patrick Schultz / Gabor Papai / Adam Ben-Shem /  Abstract: Chromatin structure is a key regulator of DNA transcription, replication and repair. In humans, the TIP60-EP400 complex (TIP60-C) is a 20-subunit assembly that affects chromatin structure through two ...Chromatin structure is a key regulator of DNA transcription, replication and repair. In humans, the TIP60-EP400 complex (TIP60-C) is a 20-subunit assembly that affects chromatin structure through two enzymatic activities: ATP-dependent exchange of histone H2A-H2B for H2A.Z-H2B, and histone acetylation. In yeast, however, these activities are performed by two independent complexes-SWR1 and NuA4, respectively. How the activities of the two complexes are merged into one supercomplex in humans, and what this association entails for the structure and mechanism of the proteins and their recruitment to chromatin, are unknown. Here we describe the structure of the endogenous human TIP60-C. We find a three-lobed architecture composed of SWR1-like (SWR1L) and NuA4-like (NuA4L) parts, which associate with a TRRAP activator-binding module. The huge EP400 subunit contains the ATPase motor, traverses the junction between SWR1L and NuA4L twice and constitutes the scaffold of the three-lobed architecture. NuA4L is completely rearranged compared with its yeast counterpart. TRRAP is flexibly tethered to NuA4L-in stark contrast to its robust connection to the completely opposite side of NuA4 in yeast. A modelled nucleosome bound to SWR1L, supported by tests of TIP60-C activity, suggests that some aspects of the histone exchange mechanism diverge from what is seen in yeast. Furthermore, a fixed actin module (as opposed to the mobile actin subcomplex in SWR1; ref. ), the flexibility of TRRAP and the weak effect of extranucleosomal DNA on exchange activity lead to a different, activator-based mode of enlisting TIP60-C to chromatin. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18611.map.gz emd_18611.map.gz | 584.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18611-v30.xml emd-18611-v30.xml emd-18611.xml emd-18611.xml | 25.9 KB 25.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_18611.png emd_18611.png | 90.7 KB | ||
| Filedesc metadata |  emd-18611.cif.gz emd-18611.cif.gz | 9.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18611 http://ftp.pdbj.org/pub/emdb/structures/EMD-18611 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18611 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18611 | HTTPS FTP |
-Validation report
| Summary document |  emd_18611_validation.pdf.gz emd_18611_validation.pdf.gz | 508.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18611_full_validation.pdf.gz emd_18611_full_validation.pdf.gz | 507.9 KB | Display | |
| Data in XML |  emd_18611_validation.xml.gz emd_18611_validation.xml.gz | 9.5 KB | Display | |
| Data in CIF |  emd_18611_validation.cif.gz emd_18611_validation.cif.gz | 11.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18611 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18611 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18611 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18611 | HTTPS FTP |
-Related structure data
| Related structure data |  8qr1MC  8qriC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18611.map.gz / Format: CCP4 / Size: 1.2 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18611.map.gz / Format: CCP4 / Size: 1.2 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map of the human Tip60 complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.73 Å | ||||||||||||||||||||||||||||||||||||
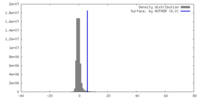
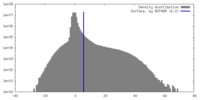
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Tip60 complex
| Entire | Name: Tip60 complex |
|---|---|
| Components |
|
-Supramolecule #1: Tip60 complex
| Supramolecule | Name: Tip60 complex / type: cell / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Strain: K562 Homo sapiens (human) / Strain: K562 |
-Macromolecule #1: E1A-binding protein p400
| Macromolecule | Name: E1A-binding protein p400 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 343.867312 KDa |
| Sequence | String: MHHGTGPQNV QHQLQRSRAC PGSEGEEQPA HPNPPPSPAA PFAPSASPSA PQSPSYQIQQ LMNRSPATGQ NVNITLQSVG PVVGGNQQI TLAPLPLPSP TSPGFQFSAQ PRRFEHGSPS YIQVTSPLSQ QVQTQSPTQP SPGPGQALQN VRAGAPGPGL G LCSSSPTG ...String: MHHGTGPQNV QHQLQRSRAC PGSEGEEQPA HPNPPPSPAA PFAPSASPSA PQSPSYQIQQ LMNRSPATGQ NVNITLQSVG PVVGGNQQI TLAPLPLPSP TSPGFQFSAQ PRRFEHGSPS YIQVTSPLSQ QVQTQSPTQP SPGPGQALQN VRAGAPGPGL G LCSSSPTG GFVDASVLVR QISLSPSSGG HFVFQDGSGL TQIAQGAQVQ LQHPGTPITV RERRPSQPHT QSGGTIHHLG PQ SPAAAGG AGLQPLASPS HITTANLPPQ ISSIIQGQLV QQQQVLQGPP LPRPLGFERT PGVLLPGAGG AAGFGMTSPP PPT SPSRTA VPPGLSSLPL TSVGNTGMKK VPKKLEEIPP ASPEMAQMRK QCLDYHYQEM QALKEVFKEY LIELFFLQHF QGNM MDFLA FKKKHYAPLQ AYLRQNDLDI EEEEEEEEEE EEKSEVINDE VKVVTGKDGQ TGTPVAIATQ LPPKVSAAFS SQQQP FQQA LAGSLVAGAG STVETDLFKR QQAMPSTGMA EQSKRPRLEV GHQGVVFQHP GADAGVPLQQ LMPTAQGGMP PTPQAA QLA GQRQSQQQYD PSTGPPVQNA ASLHTPLPQL PGRLPPAGVP TAALSSALQF AQQPQVVEAQ TQLQIPVKTQ QPNVPIP AP PSSQLPIPPS QPAQLALHVP TPGKVQVQAS QLSSLPQMVA STRLPVDPAP PCPRPLPTSS TSSLAPVSGS GPGPSPAR S SPVNRPSSAT NKALSPVTSR TPGVVASAPT KPQSPAQNAT SSQDSSQDTL TEQITLENQV HQRIAELRKA GLWSQRRLP KLQEAPRPKS HWDYLLEEMQ WMATDFAQER RWKVAAAKKL VRTVVRHHEE KQLREERGKK EEQSRLRRIA ASTAREIECF WSNIEQVVE IKLRVELEEK RKKALNLQKV SRRGKELRPK GFDALQESSL DSGMSGRKRK ASISLTDDEV DDEEETIEEE E ANEGVVDH QTELSNLAKE AELPLLDLMK LYEGAFLPSS QWPRPKPDGE DTSGEEDADD CPGDRESRKD LVLIDSLFIM DQ FKAAERM NIGKPNAKDI ADVTAVAEAI LPKGSARVTT SVKFNAPSLL YGALRDYQKI GLDWLAKLYR KNLNGILADE AGL GKTVQI IAFFAHLACN EGNWGPHLVV VRSCNILKWE LELKRWCPGL KILSYIGSHR ELKAKRQEWA EPNSFHVCIT SYTQ FFRGL TAFTRVRWKC LVIDEMQRVK GMTERHWEAV FTLQSQQRLL LIDSPLHNTF LELWTMVHFL VPGISRPYLS SPLRA PSEE SQDYYHKVVI RLHRVTQPFI LRRTKRDVEK QLTKKYEHVL KCRLSNRQKA LYEDVILQPG TQEALKSGHF VNVLSI LVR LQRICNHPGL VEPRHPGSSY VAGPLEYPSA SLILKALERD FWKEADLSMF DLIGLENKIT RHEAELLSKK KIPRKLM EE ISTSAAPAAR PAAAKLKASR LFQPVQYGQK PEGRTVAFPS THPPRTAAPT TASAAPQGPL RGRPPIATFS ANPEAKAA A APFQTSQASA SAPRHQPASA SSTAASPAHP AKLRAQTTAQ ASTPGQPPPQ PQAPSHAAGQ SALPQRLVLP SQAQARLPS GEVVKIAQLA SITGPQSRVA QPETPVTLQF QGSKFTLSHS QLRQLTAGQP LQLQGSVLQI VSAPGQPYLR APGPVVMQTV SQAGAVHGA LGSKPPAGGP SPAPLTPQVG VPGRVAVNAL AVGEPGTASK PASPIGGPTQ EEKTRLLKER LDQIYLVNER R CSQAPVYG RDLLRICALP SHGRVQWRGS LDGRRGKEAG PAHSYTSSSE SPSELMLTLC RCGESLQDVI DRVAFVIPPV VA APPSLRV PRPPPLYSHR MRILRQGLRE HAAPYFQQLR QTTAPRLLQF PELRLVQFDS GKLEALAILL QKLKSEGRRV LIL SQMILM LDILEMFLNF HYLTYVRIDE NASSEQRQEL MRSFNRDRRI FCAILSTHSR TTGINLVEAD TVVFYDNDLN PVMD AKAQE WCDRIGRCKD IHIYRLVSGN SIEEKLLKNG TKDLIREVAA QGNDYSMAFL TQRTIQELFE VYSPMDDAGF PVKAE EFVV LSQEPSVTET IAPKIARPFI EALKSIEYLE EDAQKSAQEG VLGPHTDALS SDSENMPCDE EPSQLEELAD FMEQLT PIE KYALNYLELF HTSIEQEKER NSEDAVMTAV RAWEFWNLKT LQEREARLRL EQEEAELLTY TREDAYSMEY VYEDVDG QT EVMPLWTPPT PPQDDSDIYL DSVMCLMYEA TPIPEAKLPP VYVRKERKRH KTDPSAAGRK KKQRHGEAVV PPRSLFDR A TPGLLKIRRE GKEQKKNILL KQQVPFAKPL PTFAKPTAEP GQDNPEWLIS EDWALLQAVK QLLELPLNLT IVSPAHTPN WDLVSDVVNS CSRIYRSSKQ CRNRYENVII PREEGKSKNN RPLRTSQIYA QDENATHTQL YTSHFDLMKM TAGKRSPPIK PLLGMNPFQ KNPKHASVLA ESGINYDKPL PPIQVASLRA ERIAKEKKAL ADQQKAQQPA VAQPPPPQPQ PPPPPQQPPP P LPQPQAAG SQPPAGPPAV QPQPQPQPQT QPQPVQAPAK AQPAITTGGS AAVLAGTIKT SVTGTSMPTG AVSGNVIVNT IA GVPAATF QSINKRLASP VAPGALTTPG GSAPAQVVHT QPPPRAVGSP ATATPDLVSM ATTQGVRAVT SVTASAVVTT NLT PVQTPA RSLVPQVSQA TGVQLPGKTI TPAHFQLLRQ QQQQQQQQQQ QQQQQQQQQQ QQQQQQQQTT TTSQVQVPQI QGQA QSPAQ IKAVGKLTPE HLIKMQKQKL QMPPQPPPPQ AQSAPPQPTA QVQVQTSQPP QQQSPQLTTV TAPRPGALLT GTTVA NLQV ARLTRVPTSQ LQAQGQMQTQ APQPAQVALA KPPVVSVPAA VVSSPGVTTL PMNVAGISVA IGQPQKAAGQ TVVAQP VHM QQLLKLKQQA VQQQKAIQPQ AAQGPAAVQQ KITAQQITTP GAQQKVAYAA QPALKTQFLT TPISQAQKLA GAQQVQT QI QVAKLPQVVQ QQTPVASIQQ VASASQQASP QTVALTQATA AGQQVQMIPA VTATAQVVQQ KLIQQQVVTT ASAPLQTP G APNPAQVPAS SDSPSQQPKL QMRVPAVRLK TPTKPPCQ UniProtKB: E1A-binding protein p400 |
-Macromolecule #2: Enhancer of polycomb homolog 1
| Macromolecule | Name: Enhancer of polycomb homolog 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 93.589172 KDa |
| Sequence | String: MSKLSFRARA LDASKPLPVF RCEDLPDLHE YASINRAVPQ MPTGMEKEEE SEHHLQRAIS AQQVYGEKRD NMVIPVPEAE SNIAYYESI YPGEFKMPKQ LIHIQPFSLD AEQPDYDLDS EDEVFVNKLK KKMDICPLQF EEMIDRLEKG SGQQPVSLQE A KLLLKEDD ...String: MSKLSFRARA LDASKPLPVF RCEDLPDLHE YASINRAVPQ MPTGMEKEEE SEHHLQRAIS AQQVYGEKRD NMVIPVPEAE SNIAYYESI YPGEFKMPKQ LIHIQPFSLD AEQPDYDLDS EDEVFVNKLK KKMDICPLQF EEMIDRLEKG SGQQPVSLQE A KLLLKEDD ELIREVYEYW IKKRKNCRGP SLIPSVKQEK RDGSSTNDPY VAFRRRTEKM QTRKNRKNDE ASYEKMLKLR RD LSRAVTI LEMIKRREKS KRELLHLTLE IMEKRYNLGD YNGEIMSEVM AQRQPMKPTY AIPIIPITNS SQFKHQEAMD VKE FKVNKQ DKADLIRPKR KYEKKPKVLP SSAAATPQQT SPAALPVFNA KDLNQYDFPS SDEEPLSQVL SGSSEAEEDN DPDG PFAFR RKAGCQYYAP HLDQTGNWPW TSPKDGGLGD VRYRYCLTTL TVPQRCIGFA RRRVGRGGRV LLDRAHSDYD SVFHH LDLE MLSSPQHSPV NQFANTSETN TSDKSFSKDL SQILVNIKSC RWRHFRPRTP SLHDSDNDEL SCRKLYRSIN RTGTAQ PGT QTCSTSTQSK SSSGSAHFAF TAEQYQQHQQ QLALMQKQQL AQIQQQQANS NSSTNTSQNL ASNQQKSGFR LNIQGLE RT LQGFVSKTLD SASAQFAASA LVTSEQLMGF KMKDDVVLGI GVNGVLPASG VYKGLHLSST TPTALVHTSP STAGSALL Q PSNITQTSSS HSALSHQVTA ANSATTQVLI GNNIRLTVPS SVATVNSIAP INARHIPRTL SAVPSSALKL AAAANCQVS KVPSSSSVDS VPRENHESEK PALNNIADNT VAMEVT UniProtKB: Enhancer of polycomb homolog 1 |
-Macromolecule #3: DNA methyltransferase 1-associated protein 1
| Macromolecule | Name: DNA methyltransferase 1-associated protein 1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 53.090699 KDa |
| Sequence | String: MATGADVRDI LELGGPEGDA ASGTISKKDI INPDKKKSKK SSETLTFKRP EGMHREVYAL LYSDKKDAPP LLPSDTGQGY RTVKAKLGS KKVRPWKWMP FTNPARKDGA MFFHWRRAAE EGKDYPFARF NKTVQVPVYS EQEYQLYLHD DAWTKAETDH L FDLSRRFD ...String: MATGADVRDI LELGGPEGDA ASGTISKKDI INPDKKKSKK SSETLTFKRP EGMHREVYAL LYSDKKDAPP LLPSDTGQGY RTVKAKLGS KKVRPWKWMP FTNPARKDGA MFFHWRRAAE EGKDYPFARF NKTVQVPVYS EQEYQLYLHD DAWTKAETDH L FDLSRRFD LRFVVIHDRY DHQQFKKRSV EDLKERYYHI CAKLANVRAV PGTDLKIPVF DAGHERRRKE QLERLYNRTP EQ VAEEEYL LQELRKIEAR KKEREKRSQD LQKLITAADT TAEQRRTERK APKKKLPQKK EAEKPAVPET AGIKFPDFKS AGV TLRSQR MKLPSSVGQK KIKALEQMLL ELGVELSPTP TEELVHMFNE LRSDLVLLYE LKQACANCEY ELQMLRHRHE ALAR AGVLG GPATPASGPG PASAEPAVTE PGLGPDPKDT IIDVVGAPLT PNSRKRRESA SSSSSVKKAK KP UniProtKB: DNA methyltransferase 1-associated protein 1 |
-Macromolecule #4: Vacuolar protein sorting-associated protein 72 homolog
| Macromolecule | Name: Vacuolar protein sorting-associated protein 72 homolog type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.658363 KDa |
| Sequence | String: MSLAGGRAPR KTAGNRLSGL LEAEEEDEFY QTTYGGFTEE SGDDEYQGDQ SDTEDEVDSD FDIDEGDEPS SDGEAEEPRR KRRVVTKAY KEPLKSLRPR KVNTPAGSSQ KAREEKALLP LELQDDGSDS RKSMRQSTAE HTRQTFLRVQ ERQGQSRRRK G PHCERPLT ...String: MSLAGGRAPR KTAGNRLSGL LEAEEEDEFY QTTYGGFTEE SGDDEYQGDQ SDTEDEVDSD FDIDEGDEPS SDGEAEEPRR KRRVVTKAY KEPLKSLRPR KVNTPAGSSQ KAREEKALLP LELQDDGSDS RKSMRQSTAE HTRQTFLRVQ ERQGQSRRRK G PHCERPLT QEELLREAKI TEELNLRSLE TYERLEADKK KQVHKKRKCP GPIITYHSVT VPLVGEPGPK EENVDIEGLD PA PSVSALT PHAGTGPVNP PARCSRTFIT FSDDATFEEW FPQGRPPKVP VREVCPVTHR PALYRDPVTD IPYATARAFK IIR EAYKKY ITAHGLPPTA SALGPGPPPP EPLPGSGPRA LRQKIVIK UniProtKB: Vacuolar protein sorting-associated protein 72 homolog |
-Macromolecule #5: Actin, cytoplasmic 1, N-terminally processed
| Macromolecule | Name: Actin, cytoplasmic 1, N-terminally processed / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.78266 KDa |
| Sequence | String: MDDDIAALVV DNGSGMCKAG FAGDDAPRAV FPSIVGRPRH QGVMVGMGQK DSYVGDEAQS KRGILTLKYP IEHGIVTNWD DMEKIWHHT FYNELRVAPE EHPVLLTEAP LNPKANREKM TQIMFETFNT PAMYVAIQAV LSLYASGRTT GIVMDSGDGV T HTVPIYEG ...String: MDDDIAALVV DNGSGMCKAG FAGDDAPRAV FPSIVGRPRH QGVMVGMGQK DSYVGDEAQS KRGILTLKYP IEHGIVTNWD DMEKIWHHT FYNELRVAPE EHPVLLTEAP LNPKANREKM TQIMFETFNT PAMYVAIQAV LSLYASGRTT GIVMDSGDGV T HTVPIYEG YALPHAILRL DLAGRDLTDY LMKILTERGY SFTTTAEREI VRDIKEKLCY VALDFEQEMA TAASSSSLEK SY ELPDGQV ITIGNERFRC PEALFQPSFL GMESCGIHET TFNSIMKCDV DIRKDLYANT VLSGGTTMYP GIADRMQKEI TAL APSTMK IKIIAPPERK YSVWIGGSIL ASLSTFQQMW ISKQEYDESG PSIVHRKCF UniProtKB: Actin, cytoplasmic 1 |
-Macromolecule #6: Actin-like protein 6A
| Macromolecule | Name: Actin-like protein 6A / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 47.509812 KDa |
| Sequence | String: MSGGVYGGDE VGALVFDIGS YTVRAGYAGE DCPKVDFPTA IGMVVERDDG STLMEIDGDK GKQGGPTYYI DTNALRVPRE NMEAISPLK NGMVEDWDSF QAILDHTYKM HVKSEASLHP VLMSEAPWNT RAKREKLTEL MFEHYNIPAF FLCKTAVLTA F ANGRSTGL ...String: MSGGVYGGDE VGALVFDIGS YTVRAGYAGE DCPKVDFPTA IGMVVERDDG STLMEIDGDK GKQGGPTYYI DTNALRVPRE NMEAISPLK NGMVEDWDSF QAILDHTYKM HVKSEASLHP VLMSEAPWNT RAKREKLTEL MFEHYNIPAF FLCKTAVLTA F ANGRSTGL ILDSGATHTT AIPVHDGYVL QQGIVKSPLA GDFITMQCRE LFQEMNIELV PPYMIASKEA VREGSPANWK RK EKLPQVT RSWHNYMCNC VIQDFQASVL QVSDSTYDEQ VAAQMPTVHY EFPNGYNCDF GAERLKIPEG LFDPSNVKGL SGN TMLGVS HVVTTSVGMC DIDIRPGLYG SVIVAGGNTL IQSFTDRLNR ELSQKTPPSM RLKLIANNTT VERRFSSWIG GSIL ASLGT FQQMWISKQE YEEGGKQCVE RKCP UniProtKB: Actin-like protein 6A |
-Macromolecule #7: RuvB-like 1
| Macromolecule | Name: RuvB-like 1 / type: protein_or_peptide / ID: 7 / Number of copies: 3 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.296914 KDa |
| Sequence | String: MKIEEVKSTT KTQRIASHSH VKGLGLDESG LAKQAASGLV GQENAREACG VIVELIKSKK MAGRAVLLAG PPGTGKTALA LAIAQELGS KVPFCPMVGS EVYSTEIKKT EVLMENFRRA IGLRIKETKE VYEGEVTELT PCETENPMGG YGKTISHVII G LKTAKGTK ...String: MKIEEVKSTT KTQRIASHSH VKGLGLDESG LAKQAASGLV GQENAREACG VIVELIKSKK MAGRAVLLAG PPGTGKTALA LAIAQELGS KVPFCPMVGS EVYSTEIKKT EVLMENFRRA IGLRIKETKE VYEGEVTELT PCETENPMGG YGKTISHVII G LKTAKGTK QLKLDPSIFE SLQKERVEAG DVIYIEANSG AVKRQGRCDT YATEFDLEAE EYVPLPKGDV HKKKEIIQDV TL HDLDVAN ARPQGGQDIL SMMGQLMKPK KTEITDKLRG EINKVVNKYI DQGIAELVPG VLFVDEVHML DIECFTYLHR ALE SSIAPI VIFASNRGNC VIRGTEDITS PHGIPLDLLD RVMIIRTMLY TPQEMKQIIK IRAQTEGINI SEEALNHLGE IGTK TTLRY SVQLLTPANL LAKINGKDSI EKEHVEEISE LFYDAKSSAK ILADQQDKYM K UniProtKB: RuvB-like 1 |
-Macromolecule #8: RuvB-like 2
| Macromolecule | Name: RuvB-like 2 / type: protein_or_peptide / ID: 8 / Number of copies: 3 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 51.222465 KDa |
| Sequence | String: MATVTATTKV PEIRDVTRIE RIGAHSHIRG LGLDDALEPR QASQGMVGQL AARRAAGVVL EMIREGKIAG RAVLIAGQPG TGKTAIAMG MAQALGPDTP FTAIAGSEIF SLEMSKTEAL TQAFRRSIGV RIKEETEIIE GEVVEIQIDR PATGTGSKVG K LTLKTTEM ...String: MATVTATTKV PEIRDVTRIE RIGAHSHIRG LGLDDALEPR QASQGMVGQL AARRAAGVVL EMIREGKIAG RAVLIAGQPG TGKTAIAMG MAQALGPDTP FTAIAGSEIF SLEMSKTEAL TQAFRRSIGV RIKEETEIIE GEVVEIQIDR PATGTGSKVG K LTLKTTEM ETIYDLGTKM IESLTKDKVQ AGDVITIDKA TGKISKLGRS FTRARDYDAM GSQTKFVQCP DGELQKRKEV VH TVSLHEI DVINSRTQGF LALFSGDTGE IKSEVREQIN AKVAEWREEG KAEIIPGVLF IDEVHMLDIE SFSFLNRALE SDM APVLIM ATNRGITRIR GTSYQSPHGI PIDLLDRLLI VSTTPYSEKD TKQILRIRCE EEDVEMSEDA YTVLTRIGLE TSLR YAIQL ITAASLVCRK RKGTEVQVDD IKRVYSLFLD ESRSTQYMKE YQDAFLFNEL KGETMDTS UniProtKB: RuvB-like 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 279 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: FEI FALCON IV (4k x 4k) / #0 - Average electron dose: 40.0 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / #1 - Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8qr1: |
 Movie
Movie Controller
Controller









































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)