+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mouse leptin:LEP-R complex cryoEM structure (3:3 model) | |||||||||
 Map data Map data | Sharpenend cryo-EM map following non-uniform refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | leptin / LEP-R / obesity / metabolism / energy balance / CYTOKINE | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of metabolic process / negative regulation of locomotor rhythm / negative regulation of eating behavior / Synthesis, secretion, and deacylation of Ghrelin / regulation of lipoprotein lipid oxidation / cellular response to L-ascorbic acid / positive regulation of fat cell apoptotic process / negative regulation of glutamine transport / leptin receptor activity / negative regulation of appetite by leptin-mediated signaling pathway ...negative regulation of metabolic process / negative regulation of locomotor rhythm / negative regulation of eating behavior / Synthesis, secretion, and deacylation of Ghrelin / regulation of lipoprotein lipid oxidation / cellular response to L-ascorbic acid / positive regulation of fat cell apoptotic process / negative regulation of glutamine transport / leptin receptor activity / negative regulation of appetite by leptin-mediated signaling pathway / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / negative regulation of glucagon secretion / regulation of endothelial cell proliferation / regulation of natural killer cell proliferation / leptin receptor binding / regulation of natural killer cell mediated cytotoxicity / protein-hormone receptor activity / bone growth / positive regulation of luteinizing hormone secretion / regulation of natural killer cell activation / glycerol biosynthetic process / positive regulation of monoatomic ion transport / elastin metabolic process / leptin-mediated signaling pathway / positive regulation of follicle-stimulating hormone secretion / regulation of steroid biosynthetic process / regulation of intestinal cholesterol absorption / regulation of bone remodeling / regulation of brown fat cell differentiation / positive regulation of peroxisome proliferator activated receptor signaling pathway / positive regulation of hepatic stellate cell activation / adult feeding behavior / regulation of nitric-oxide synthase activity / response to leptin / regulation of lipid biosynthetic process / bone mineralization involved in bone maturation / regulation of feeding behavior / sexual reproduction / activation of protein kinase C activity / negative regulation of cartilage development / fatty acid catabolic process / ovulation from ovarian follicle / negative regulation of appetite / positive regulation of developmental growth / leukocyte tethering or rolling / cellular response to leptin stimulus / energy reserve metabolic process / bile acid metabolic process / negative regulation of D-glucose import / tyrosine phosphorylation of STAT protein / prostaglandin secretion / regulation of protein localization to nucleus / cardiac muscle hypertrophy / hormone metabolic process / regulation of metabolic process / aorta development / intestinal absorption / insulin secretion / cytokine receptor activity / regulation of fat cell differentiation / positive regulation of p38MAPK cascade / negative regulation of vasoconstriction / eating behavior / peptide hormone receptor binding / regulation of gluconeogenesis / glycogen metabolic process / fatty acid beta-oxidation / regulation of cytokine production involved in inflammatory response / cytokine binding / central nervous system neuron development / positive regulation of insulin secretion involved in cellular response to glucose stimulus / peptide hormone binding / response to dietary excess / negative regulation of lipid storage / T cell differentiation / positive regulation of TOR signaling / response to vitamin E / glial cell proliferation / negative regulation of gluconeogenesis / adipose tissue development / regulation of angiogenesis / phagocytosis / positive regulation of insulin receptor signaling pathway / positive regulation of T cell proliferation / cellular response to retinoic acid / positive regulation of tyrosine phosphorylation of STAT protein / energy homeostasis / positive regulation of interleukin-12 production / cholesterol metabolic process / regulation of insulin secretion / negative regulation of autophagy / determination of adult lifespan / response to activity / gluconeogenesis / positive regulation of interleukin-8 production / female pregnancy / positive regulation of receptor signaling pathway via JAK-STAT / response to insulin / regulation of protein phosphorylation / placenta development Similarity search - Function | |||||||||
| Biological species |  | |||||||||
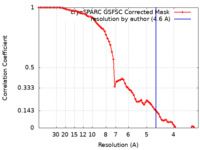
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||
 Authors Authors | Verstraete K / Savvides SN / Verschueren KG / Tsirigotaki A | |||||||||
| Funding support |  Belgium, 1 items Belgium, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Mechanism of receptor assembly via the pleiotropic adipokine Leptin. Authors: Alexandra Tsirigotaki / Ann Dansercoer / Koen H G Verschueren / Iva Marković / Christoph Pollmann / Maximillian Hafer / Jan Felix / Catherine Birck / Wouter Van Putte / Dominiek Catteeuw / ...Authors: Alexandra Tsirigotaki / Ann Dansercoer / Koen H G Verschueren / Iva Marković / Christoph Pollmann / Maximillian Hafer / Jan Felix / Catherine Birck / Wouter Van Putte / Dominiek Catteeuw / Jan Tavernier / J Fernando Bazan / Jacob Piehler / Savvas N Savvides / Kenneth Verstraete /     Abstract: The adipokine Leptin activates its receptor LEP-R in the hypothalamus to regulate body weight and exerts additional pleiotropic functions in immunity, fertility and cancer. However, the structure and ...The adipokine Leptin activates its receptor LEP-R in the hypothalamus to regulate body weight and exerts additional pleiotropic functions in immunity, fertility and cancer. However, the structure and mechanism of Leptin-mediated LEP-R assemblies has remained unclear. Intriguingly, the signaling-competent isoform of LEP-R is only lowly abundant amid several inactive short LEP-R isoforms contributing to a mechanistic conundrum. Here we show by X-ray crystallography and cryo-EM that, in contrast to long-standing paradigms, Leptin induces type I cytokine receptor assemblies featuring 3:3 stoichiometry and demonstrate such Leptin-induced trimerization of LEP-R on living cells via single-molecule microscopy. In mediating these assemblies, Leptin undergoes drastic restructuring that activates its site III for binding to the Ig domain of an adjacent LEP-R. These interactions are abolished by mutations linked to obesity. Collectively, our study provides the structural and mechanistic framework for how evolutionarily conserved Leptin:LEP-R assemblies with 3:3 stoichiometry can engage distinct LEP-R isoforms to achieve signaling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15678.map.gz emd_15678.map.gz | 40.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15678-v30.xml emd-15678-v30.xml emd-15678.xml emd-15678.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15678_fsc.xml emd_15678_fsc.xml | 7.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_15678.png emd_15678.png | 117.4 KB | ||
| Masks |  emd_15678_msk_1.map emd_15678_msk_1.map | 42.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15678.cif.gz emd-15678.cif.gz | 7.2 KB | ||
| Others |  emd_15678_additional_1.map.gz emd_15678_additional_1.map.gz emd_15678_half_map_1.map.gz emd_15678_half_map_1.map.gz emd_15678_half_map_2.map.gz emd_15678_half_map_2.map.gz | 21.5 MB 39.8 MB 39.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15678 http://ftp.pdbj.org/pub/emdb/structures/EMD-15678 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15678 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15678 | HTTPS FTP |
-Validation report
| Summary document |  emd_15678_validation.pdf.gz emd_15678_validation.pdf.gz | 859.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15678_full_validation.pdf.gz emd_15678_full_validation.pdf.gz | 858.7 KB | Display | |
| Data in XML |  emd_15678_validation.xml.gz emd_15678_validation.xml.gz | 14.7 KB | Display | |
| Data in CIF |  emd_15678_validation.cif.gz emd_15678_validation.cif.gz | 18.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15678 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15678 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15678 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15678 | HTTPS FTP |
-Related structure data
| Related structure data |  8avcMC  7z3pC  7z3qC  7z3rC  8av2C  8avbC  8avdC  8aveC  8avfC  8avoC  8b7qC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15678.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15678.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpenend cryo-EM map following non-uniform refinement | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.658 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15678_msk_1.map emd_15678_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Non-sharpened cryo-EM map following non-uniform refinement
| File | emd_15678_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Non-sharpened cryo-EM map following non-uniform refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_15678_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_15678_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Stucture of mouse leptin in complex with a trimerized form of the...
| Entire | Name: Stucture of mouse leptin in complex with a trimerized form of the mouse Lep-R extracellular region |
|---|---|
| Components |
|
-Supramolecule #1: Stucture of mouse leptin in complex with a trimerized form of the...
| Supramolecule | Name: Stucture of mouse leptin in complex with a trimerized form of the mouse Lep-R extracellular region type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: The mLEP-R ectodomain was C-terminally fused to a trimeric GCN4 isoleucine zipper tag and secreted from HEK93 FreeStyle cells and complex with refolded mouse leptin produced in E.coli. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 444 KDa |
-Macromolecule #1: Leptin
| Macromolecule | Name: Leptin / type: protein_or_peptide / ID: 1 Details: N-terminally His-tagged mouse leptin was refolded from inclusion bodies produced in E.coli. Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.873283 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH PGGPGSENLY FQGGSTGGVP IQKVQDDTKT LIKTIVTRIN DISHTQSVSA KQRVTGLDFI PGLHPILSLS KMDQTLAVY QQVLTSLPSQ NVLQIANDLE NLRDLLHLLA FSKSCSLPQT SGLQKPESLD GVLEASLYST EVVALSRLQG S LQDILQQL DVSPEC UniProtKB: Leptin |
-Macromolecule #2: Leptin receptor
| Macromolecule | Name: Leptin receptor / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 97.479391 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LNLAYPISPW KFKLFCGPPN TTDDSFLSPA GAPNNASALK GASEAIVEAK FNSSGIYVPE LSKTVFHCCF GNEQGQNCSA LTDNTEGKT LASVVKASVF RQLGVNWDIE CWMKGDLTLF ICHMEPLPKN PFKNYDSKVH LLYDLPEVID DSPLPPLKDS F QTVQCNCS ...String: LNLAYPISPW KFKLFCGPPN TTDDSFLSPA GAPNNASALK GASEAIVEAK FNSSGIYVPE LSKTVFHCCF GNEQGQNCSA LTDNTEGKT LASVVKASVF RQLGVNWDIE CWMKGDLTLF ICHMEPLPKN PFKNYDSKVH LLYDLPEVID DSPLPPLKDS F QTVQCNCS LRGCECHVPV PRAKLNYALL MYLEITSAGV SFQSPLMSLQ PMLVVKPDPP LGLHMEVTDD GNLKISWDSQ TM APFPLQY QVKYLENSTI VREAAEIVSA TSLLVDSVLP GSSYEVQVRS KRLDGSGVWS DWSSPQVFTT QDVVYFPPKI LTS VGSNAS FHCIYKNENQ IISSKQIVWW RNLAEKIPEI QYSIVSDRVS KVTFSNLKAT RPRGKFTYDA VYCCNEQACH HRYA ELYVI DVNINISCET DGYLTKMTCR WSPSTIQSLV GSTVQLRYHR RSLYCPDSPS IHPTSEPKNC VLQRDGFYEC VFQPI FLLS GYTMWIRINH SLGSLDSPPT CVLPDSVVKP LPPSNVKAEI TVNTGLLKVS WEKPVFPENN LQFQIRYGLS GKEIQW KTH EVFDAKSKSA SLLVSDLCAV YVVQVRCRRL DGLGYWSNWS SPAYTLVMDV KVPMRGPEFW RKMDGDVTKK ERNVTLL WK PLTKNDSLCS VRRYVVKHRT AHNGTWSEDV GNRTNLTFLW TEPAHTVTVL AVNSLGASLV NFNLTFSWPM SKVSAVES L SAYPLSSSCV ILSWTLSPDD YSLLYLVIEW KILNEDDGMK WLRIPSNVKK FYIHDNFIPI EKYQFSLYPV FMEGVGKPK IINGFTKDAI DKQQNDAGST GGSGGSGGSG GSGGSRMKQI EDKIEEILSK IYHIENEIAR IKKLIGER UniProtKB: Leptin receptor |
-Macromolecule #3: NICKEL (II) ION
| Macromolecule | Name: NICKEL (II) ION / type: ligand / ID: 3 / Number of copies: 3 / Formula: NI |
|---|---|
| Molecular weight | Theoretical: 58.693 Da |
| Chemical component information |  ChemComp-NI: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: 20 mM HEPES, 150 mM NaCl, pH 7.4 | |||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 99 % / Chamber temperature: 22 K / Instrument: LEICA EM GP | |||||||||
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 13230 / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8avc: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)