+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human TREX core THO-UAP56 complex (map D) | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transcription and export complex / TREX / RNA export / RNA packaging / RNA binding protein / gene expression / GENE REGULATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationTHO complex / THO complex part of transcription export complex / transcription export complex / primitive hemopoiesis / regulation of mRNA export from nucleus / U6 snRNP / RNA secondary structure unwinding / stem cell division / ATP-dependent protein binding / mRNA 3'-end processing ...THO complex / THO complex part of transcription export complex / transcription export complex / primitive hemopoiesis / regulation of mRNA export from nucleus / U6 snRNP / RNA secondary structure unwinding / stem cell division / ATP-dependent protein binding / mRNA 3'-end processing / ATP-dependent activity, acting on RNA / U4 snRNA binding / RNA export from nucleus / Transport of Mature mRNA derived from an Intron-Containing Transcript / U4 snRNP / RNA Polymerase II Transcription Termination / poly(A)+ mRNA export from nucleus / generation of neurons / spliceosomal complex assembly / monocyte differentiation / blastocyst development / RHOBTB2 GTPase cycle / mRNA export from nucleus / neuron development / U6 snRNA binding / mRNA Splicing - Major Pathway / RNA splicing / central nervous system development / spliceosomal complex / cell morphogenesis / mRNA splicing, via spliceosome / nuclear matrix / mRNA processing / Signaling by CSF1 (M-CSF) in myeloid cells / negative regulation of neuron projection development / regulation of gene expression / RNA helicase activity / nuclear body / RNA helicase / nuclear speck / mRNA binding / apoptotic process / signal transduction / ATP hydrolysis activity / DNA binding / RNA binding / nucleoplasm / ATP binding / identical protein binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
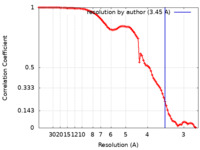
| Method | single particle reconstruction / cryo EM / Resolution: 3.45 Å | |||||||||
 Authors Authors | Pacheco-Fiallos FB / Vorlaender MK / Plaschka C | |||||||||
| Funding support | European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: mRNA recognition and packaging by the human transcription-export complex. Authors: Belén Pacheco-Fiallos / Matthias K Vorländer / Daria Riabov-Bassat / Laura Fin / Francis J O'Reilly / Farja I Ayala / Ulla Schellhaas / Juri Rappsilber / Clemens Plaschka /    Abstract: Newly made mRNAs are processed and packaged into mature ribonucleoprotein complexes (mRNPs) and are recognized by the essential transcription-export complex (TREX) for nuclear export. However, the ...Newly made mRNAs are processed and packaged into mature ribonucleoprotein complexes (mRNPs) and are recognized by the essential transcription-export complex (TREX) for nuclear export. However, the mechanisms of mRNP recognition and three-dimensional mRNP organization are poorly understood. Here we report cryo-electron microscopy and tomography structures of reconstituted and endogenous human mRNPs bound to the 2-MDa TREX complex. We show that mRNPs are recognized through multivalent interactions between the TREX subunit ALYREF and mRNP-bound exon junction complexes. Exon junction complexes can multimerize through ALYREF, which suggests a mechanism for mRNP organization. Endogenous mRNPs form compact globules that are coated by multiple TREX complexes. These results reveal how TREX may simultaneously recognize, compact and protect mRNAs to promote their packaging for nuclear export. The organization of mRNP globules provides a framework to understand how mRNP architecture facilitates mRNA biogenesis and export. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14808.map.gz emd_14808.map.gz | 60.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14808-v30.xml emd-14808-v30.xml emd-14808.xml emd-14808.xml | 27.9 KB 27.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14808_fsc.xml emd_14808_fsc.xml | 8.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_14808.png emd_14808.png | 48.1 KB | ||
| Filedesc metadata |  emd-14808.cif.gz emd-14808.cif.gz | 8.7 KB | ||
| Others |  emd_14808_additional_1.map.gz emd_14808_additional_1.map.gz emd_14808_half_map_1.map.gz emd_14808_half_map_1.map.gz emd_14808_half_map_2.map.gz emd_14808_half_map_2.map.gz | 31.3 MB 59.1 MB 59.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14808 http://ftp.pdbj.org/pub/emdb/structures/EMD-14808 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14808 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14808 | HTTPS FTP |
-Validation report
| Summary document |  emd_14808_validation.pdf.gz emd_14808_validation.pdf.gz | 545.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14808_full_validation.pdf.gz emd_14808_full_validation.pdf.gz | 545.3 KB | Display | |
| Data in XML |  emd_14808_validation.xml.gz emd_14808_validation.xml.gz | 21 KB | Display | |
| Data in CIF |  emd_14808_validation.cif.gz emd_14808_validation.cif.gz | 26.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14808 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14808 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14808 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14808 | HTTPS FTP |
-Related structure data
| Related structure data |  7znlMC  7znjC  7znkC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14808.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14808.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened map
| File | emd_14808_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
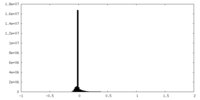
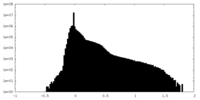
| Density Histograms |
-Half map: Half map 1
| File | emd_14808_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
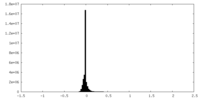
| Density Histograms |
-Half map: Half map 2
| File | emd_14808_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
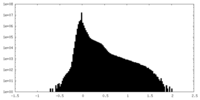
| Density Histograms |
- Sample components
Sample components
-Entire : THO-UAP56
| Entire | Name: THO-UAP56 |
|---|---|
| Components |
|
-Supramolecule #1: THO-UAP56
| Supramolecule | Name: THO-UAP56 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.8 MDa |
-Macromolecule #1: THO complex subunit 1
| Macromolecule | Name: THO complex subunit 1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 75.752156 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSPTPPLFSL PEARTRFTKS TREALNNKNI KPLLSTFSQV PGSENEKKCT LDQAFRGILE EEIINHSSCE NVLAIISLAI GGVTEGICT ASTPFVLLGD VLDCLPLDQC DTIFTFVEKN VATWKSNTFY SAGKNYLLRM CNDLLRRLSK SQNTVFCGRI Q LFLARLFP ...String: MSPTPPLFSL PEARTRFTKS TREALNNKNI KPLLSTFSQV PGSENEKKCT LDQAFRGILE EEIINHSSCE NVLAIISLAI GGVTEGICT ASTPFVLLGD VLDCLPLDQC DTIFTFVEKN VATWKSNTFY SAGKNYLLRM CNDLLRRLSK SQNTVFCGRI Q LFLARLFP LSEKSGLNLQ SQFNLENVTV FNTNEQESTL GQKHTEDREE GMDVEEGEMG DEEAPTTCSI PIDYNLYRKF WS LQDYFRN PVQCYEKISW KTFLKYSEEV LAVFKSYKLD DTQASRKKME ELKTGGEHVY FAKFLTSEKL MDLQLSDSNF RRH ILLQYL ILFQYLKGQV KFKSSNYVLT DEQSLWIEDT TKSVYQLLSE NPPDGERFSK MVEHILNTEE NWNSWKNEGC PSFV KERTS DTKPTRIIRK RTAPEDFLGK GPTKKILMGN EELTRLWNLC PDNMEACKSE TREHMPTLEE FFEEAIEQAD PENMV ENEY KAVNNSNYGW RALRLLARRS PHFFQPTNQQ FKSLPEYLEN MVIKLAKELP PPSEEIKTGE DEDEEDNDAL LKENES PDV RRDKPVTGEQ IEVFANKLGE QWKILAPYLE MKDSEIRQIE CDSEDMKMRA KQLLVAWQDQ EGVHATPENL INALNKS GL SDLAESLTND NETNS UniProtKB: THO complex subunit 1 |
-Macromolecule #2: THO complex subunit 2
| Macromolecule | Name: THO complex subunit 2 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 183.087734 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAAAAVVVPA EWIKNWEKSG RGEFLHLCRI LSENKSHDSS TYRDFQQALY ELSYHVIKGN LKHEQASNVL SDISEFREDM PSILADVFC ILDIETNCLE EKSKRDYFTQ LVLACLYLVS DTVLKERLDP ETLESLGLIK QSQQFNQKSV KIKTKLFYKQ Q KFNLLREE ...String: MAAAAVVVPA EWIKNWEKSG RGEFLHLCRI LSENKSHDSS TYRDFQQALY ELSYHVIKGN LKHEQASNVL SDISEFREDM PSILADVFC ILDIETNCLE EKSKRDYFTQ LVLACLYLVS DTVLKERLDP ETLESLGLIK QSQQFNQKSV KIKTKLFYKQ Q KFNLLREE NEGYAKLIAE LGQDLSGSIT SDLILENIKS LIGCFNLDPN RVLDVILEVF ECRPEHDDFF ISLLESYMSM CE PQTLCHI LGFKFKFYQE PNGETPSSLY RVAAVLLQFN LIDLDDLYVH LLPADNCIMD EHKREIAEAK QIVRKLTMVV LSS EKMDER EKEKEKEEEK VEKPPDNQKL GLLEALLKIG DWQHAQNIMD QMPPYYAASH KLIALAICKL IHITIEPLYR RVGV PKGAK GSPVNALQNK RAPKQAESFE DLRRDVFNMF CYLGPHLSHD PILFAKVVRI GKSFMKEFQS DGSKQEDKEK TEVIL SCLL SITDQVLLPS LSLMDCNACM SEELWGMFKT FPYQHRYRLY GQWKNETYNS HPLLVKVKAQ TIDRAKYIMK RLTKEN VKP SGRQIGKLSH SNPTILFDYI LSQIQKYDNL ITPVVDSLKY LTSLNYDVLA YCIIEALANP EKERMKHDDT TISSWLQ SL ASFCGAVFRK YPIDLAGLLQ YVANQLKAGK SFDLLILKEV VQKMAGIEIT EEMTMEQLEA MTGGEQLKAE GGYFGQIR N TKKSSQRLKD ALLDHDLALP LCLLMAQQRN GVIFQEGGEK HLKLVGKLYD QCHDTLVQFG GFLASNLSTE DYIKRVPSI DVLCNEFHTP HDAAFFLSRP MYAHHISSKY DELKKSEKGS KQQHKVHKYI TSCEMVMAPV HEAVVSLHVS KVWDDISPQF YATFWSLTM YDLAVPHTSY EREVNKLKVQ MKAIDDNQEM PPNKKKKEKE RCTALQDKLL EEEKKQMEHV QRVLQRLKLE K DNWLLAKS TKNETITKFL QLCIFPRCIF SAIDAVYCAR FVELVHQQKT PNFSTLLCYD RVFSDIIYTV ASCTENEASR YG RFLCCML ETVTRWHSDR ATYEKECGNY PGFLTILRAT GFDGGNKADQ LDYENFRHVV HKWHYKLTKA SVHCLETGEY THI RNILIV LTKILPWYPK VLNLGQALER RVHKICQEEK EKRPDLYALA MGYSGQLKSR KSYMIPENEF HHKDPPPRNA VASV QNGPG GGPSSSSIGS ASKSDESSTE ETDKSRERSQ CGVKAVNKAS STTPKGNSSN GNSGSNSNKA VKENDKEKGK EKEKE KKEK TPATTPEARV LGKDGKEKPK EERPNKDEKA RETKERTPKS DKEKEKFKKE EKAKDEKFKT TVPNAESKST QERERE KEP SRERDIAKEM KSKENVKGGE KTPVSGSLKS PVPRSDIPEP EREQKRRKID THPSPSHSST VKDSLIELKE SSAKLYI NH TPPPLSKSKE REMDKKDLDK SRERSREREK KDEKDRKERK RDHSNNDREV PPDLTKRRKE ENGTMGVSKH KSESPCES P YPNEKDKEKN KSKSSGKEKG SDSFKSEKMD KISSGGKKES RHDKEKIEKK EKRDSSGGKE EKKHHKSSDK HR UniProtKB: THO complex subunit 2 |
-Macromolecule #3: THO complex subunit 3
| Macromolecule | Name: THO complex subunit 3 / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.817617 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAVPAAAMGP SALGQSGPGS MAPWCSVSSG PSRYVLGMQE LFRGHSKTRE FLAHSAKVHS VAWSCDGRRL ASGSFDKTAS VFLLEKDRL VKENNYRGHG DSVDQLCWHP SNPDLFVTAS GDKTIRIWDV RTTKCIATVN TKGENINICW SPDGQTIAVG N KDDVVTFI ...String: MAVPAAAMGP SALGQSGPGS MAPWCSVSSG PSRYVLGMQE LFRGHSKTRE FLAHSAKVHS VAWSCDGRRL ASGSFDKTAS VFLLEKDRL VKENNYRGHG DSVDQLCWHP SNPDLFVTAS GDKTIRIWDV RTTKCIATVN TKGENINICW SPDGQTIAVG N KDDVVTFI DAKTHRSKAE EQFKFEVNEI SWNNDNNMFF LTNGNGCINI LSYPELKPVQ SINAHPSNCI CIKFDPMGKY FA TGSADAL VSLWDVDELV CVRCFSRLDW PVRTLSFSHD GKMLASASED HFIDIAEVET GDKLWEVQCE SPTFTVAWHP KRP LLAFAC DDKDGKYDSS REAGTVKLFG LPNDS UniProtKB: THO complex subunit 3 |
-Macromolecule #4: THO complex subunit 5 homolog
| Macromolecule | Name: THO complex subunit 5 homolog / type: protein_or_peptide / ID: 4 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 78.624852 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSSESSKKRK PKVIRSDGAP AEGKRNRSDT EQEGKYYSEE AEVDLRDPGR DYELYKYTCQ ELQRLMAEIQ DLKSRGGKDV AIEIEERRI QSCVHFMTLK KLNRLAHIRL KKGRDQTHEA KQKVDAYHLQ LQNLLYEVMH LQKEITKCLE FKSKHEEIDL V SLEEFYKE ...String: MSSESSKKRK PKVIRSDGAP AEGKRNRSDT EQEGKYYSEE AEVDLRDPGR DYELYKYTCQ ELQRLMAEIQ DLKSRGGKDV AIEIEERRI QSCVHFMTLK KLNRLAHIRL KKGRDQTHEA KQKVDAYHLQ LQNLLYEVMH LQKEITKCLE FKSKHEEIDL V SLEEFYKE APPDISKAEV TMGDPHQQTL ARLDWELEQR KRLAEKYREC LSNKEKILKE IEVKKEYLSS LQPRLNSIMQ AS LPVQEYL FMPFDQAHKQ YETARHLPPP LYVLFVQATA YGQACDKTLS VAIEGSVDEA KALFKPPEDS QDDESDSDAE EEQ TTKRRR PTLGVQLDDK RKEMLKRHPL SVMLDLKCKD DSVLHLTFYY LMNLNIMTVK AKVTTAMELI TPISAGDLLS PDSV LSCLY PGDHGKKTPN PANQYQFDKV GILTLSDYVL ELGHPYLWVQ KLGGLHFPKE QPQQTVIADH SLSASHMETT MKLLK TRVQ SRLALHKQFA SLEHGIVPVT SDCQYLFPAK VVSRLVKWVT VAHEDYMELH FTKDIVDAGL AGDTNLYYMA LIERGT AKL QAAVVLNPGY SSIPPVFQLC LNWKGEKTNS NDDNIRAMEG EVNVCYKELC GPWPSHQLLT NQLQRLCVLL DVYLETE SH DDSVEGPKEF PQEKMCLRLF RGPSRMKPFK YNHPQGFFSH R UniProtKB: THO complex subunit 5 |
-Macromolecule #5: THO complex subunit 6 homolog
| Macromolecule | Name: THO complex subunit 6 homolog / type: protein_or_peptide / ID: 5 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.577875 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MERAVPLAVP LGQTEVFQAL QRLHMTIFSQ SVSPCGKFLA AGNNYGQIAI FSLSSALSSE AKEESKKPVV TFQAHDGPVY SMVSTDRHL LSAGDGEVKA WLWAEMLKKG CKELWRRQPP YRTSLEVPEI NALLLVPKEN SLILAGGDCQ LHTMDLETGT F TRVLRGHT ...String: MERAVPLAVP LGQTEVFQAL QRLHMTIFSQ SVSPCGKFLA AGNNYGQIAI FSLSSALSSE AKEESKKPVV TFQAHDGPVY SMVSTDRHL LSAGDGEVKA WLWAEMLKKG CKELWRRQPP YRTSLEVPEI NALLLVPKEN SLILAGGDCQ LHTMDLETGT F TRVLRGHT DYIHCLALRE RSPEVLSGGE DGAVRLWDLR TAKEVQTIEV YKHEECSRPH NGRWIGCLAT DSDWMVCGGG PA LTLWHLR SSTPTTIFPI RAPQKHVTFY QDLILSAGQG RCVNQWQLSG ELKAQVPGSS PGLLSLSLNQ QPAAPECKVL TAA GNSCRV DVFTNLGYRA FSLSF UniProtKB: THO complex subunit 6 |
-Macromolecule #6: THO complex subunit 7 homolog
| Macromolecule | Name: THO complex subunit 7 homolog / type: protein_or_peptide / ID: 6 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.782014 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGAVTDDEVI RKRLLIDGDG AGDDRRINLL VKSFIKWCNS GSQEEGYSQY QRMLSTLSQC EFSMGKTLLV YDMNLREMEN YEKIYKEIE CSIAGAHEKI AECKKQILQA KRIRKNRQEY DALAKVIQHH PDRHETLKEL EALGKELEHL SHIKESVEDK L ELRRKQFH ...String: MGAVTDDEVI RKRLLIDGDG AGDDRRINLL VKSFIKWCNS GSQEEGYSQY QRMLSTLSQC EFSMGKTLLV YDMNLREMEN YEKIYKEIE CSIAGAHEKI AECKKQILQA KRIRKNRQEY DALAKVIQHH PDRHETLKEL EALGKELEHL SHIKESVEDK L ELRRKQFH VLLSTIHELQ QTLENDEKLS EVEEAQEASM ETDPKP UniProtKB: THO complex subunit 7 |
-Macromolecule #7: Spliceosome RNA helicase DDX39B
| Macromolecule | Name: Spliceosome RNA helicase DDX39B / type: protein_or_peptide / ID: 7 / Number of copies: 4 / Enantiomer: LEVO / EC number: RNA helicase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 49.05625 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAENDVDNEL LDYEDDEVET AAGGDGAEAP AKKDVKGSYV SIHSSGFRDF LLKPELLRAI VDCGFEHPSE VQHECIPQAI LGMDVLCQA KSGMGKTAVF VLATLQQLEP VTGQVSVLVM CHTRELAFQI SKEYERFSKY MPNVKVAVFF GGLSIKKDEE V LKKNCPHI ...String: MAENDVDNEL LDYEDDEVET AAGGDGAEAP AKKDVKGSYV SIHSSGFRDF LLKPELLRAI VDCGFEHPSE VQHECIPQAI LGMDVLCQA KSGMGKTAVF VLATLQQLEP VTGQVSVLVM CHTRELAFQI SKEYERFSKY MPNVKVAVFF GGLSIKKDEE V LKKNCPHI VVGTPGRILA LARNKSLNLK HIKHFILDEC DKMLEQLDMR RDVQEIFRMT PHEKQVMMFS ATLSKEIRPV CR KFMQDPM EIFVDDETKL TLHGLQQYYV KLKDNEKNRK LFDLLDVLEF NQVVIFVKSV QRCIALAQLL VEQNFPAIAI HRG MPQEER LSRYQQFKDF QRRILVATNL FGRGMDIERV NIAFNYDMPE DSDTYLHRVA RAGRFGTKGL AITFVSDEND AKIL NDVQD RFEVNISELP DEIDISSYIE QTR UniProtKB: Spliceosome RNA helicase DDX39B |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.7 µm / Nominal defocus min: 0.4 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)