[日本語] English
 万見
万見- EMDB-12721: Yeast TFIIH in the contracted state within the pre-initiation complex -
+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-12721 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Yeast TFIIH in the contracted state within the pre-initiation complex | ||||||||||||||||||
 マップデータ マップデータ | |||||||||||||||||||
 試料 試料 |
| ||||||||||||||||||
 キーワード キーワード | Pre-initiation complex / TRANSCRIPTION | ||||||||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報regulation of mitotic recombination / RNA polymerase II promoter clearance / phosphatidylinositol-5-phosphate binding / positive regulation of mitotic recombination / nucleotide-excision repair factor 3 complex / transcription factor TFIIE complex / DNA translocase activity / nucleotide-excision repair, preincision complex assembly / transcription factor TFIIK complex / transcription open complex formation at RNA polymerase II promoter ...regulation of mitotic recombination / RNA polymerase II promoter clearance / phosphatidylinositol-5-phosphate binding / positive regulation of mitotic recombination / nucleotide-excision repair factor 3 complex / transcription factor TFIIE complex / DNA translocase activity / nucleotide-excision repair, preincision complex assembly / transcription factor TFIIK complex / transcription open complex formation at RNA polymerase II promoter / transcriptional start site selection at RNA polymerase II promoter / RPB4-RPB7 complex / DNA 5'-3' helicase / phosphatidylinositol-3-phosphate binding / transcription factor TFIIH holo complex / transcription factor TFIIH core complex / cyclin-dependent protein serine/threonine kinase activator activity / DNA 3'-5' helicase / transcription preinitiation complex / poly(A)+ mRNA export from nucleus / RNA Polymerase I Transcription Initiation / DNA duplex unwinding / Processing of Capped Intron-Containing Pre-mRNA / nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay / RNA Polymerase III Transcription Initiation From Type 2 Promoter / 3'-5' DNA helicase activity / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / mRNA Capping / RNA polymerase II transcribes snRNA genes / TP53 Regulates Transcription of DNA Repair Genes / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / RNA Polymerase II Pre-transcription Events / RNA-templated transcription / termination of RNA polymerase III transcription / Formation of TC-NER Pre-Incision Complex / transcription initiation at RNA polymerase III promoter / termination of RNA polymerase I transcription / RNA Polymerase I Promoter Escape / RNA polymerase II complex binding / nucleolar large rRNA transcription by RNA polymerase I / Gap-filling DNA repair synthesis and ligation in TC-NER / transcription by RNA polymerase I / transcription initiation at RNA polymerase I promoter / nuclear-transcribed mRNA catabolic process / ATPase activator activity / Estrogen-dependent gene expression / positive regulation of nuclear-transcribed mRNA poly(A) tail shortening / transcription by RNA polymerase III / Dual incision in TC-NER / transcription elongation by RNA polymerase I / tRNA transcription by RNA polymerase III / RNA polymerase I complex / RNA polymerase I activity / RNA polymerase III complex / translesion synthesis / positive regulation of translational initiation / ATP-dependent activity, acting on DNA / RNA polymerase II, core complex / translation initiation factor binding / DNA helicase activity / isomerase activity / DNA-templated transcription initiation / transcription initiation at RNA polymerase II promoter / nucleotide-excision repair / transcription elongation by RNA polymerase II / P-body / ribonucleoside binding / DNA-directed 5'-3' RNA polymerase activity / DNA-directed RNA polymerase / cytoplasmic stress granule / mRNA processing / ubiquitin protein ligase activity / ribosome biogenesis / single-stranded DNA binding / 4 iron, 4 sulfur cluster binding / 5'-3' DNA helicase activity / double-stranded DNA binding / transcription by RNA polymerase II / damaged DNA binding / single-stranded RNA binding / DNA repair / mRNA binding / nucleotide binding / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / ATP hydrolysis activity / mitochondrion / DNA binding / zinc ion binding / nucleoplasm / ATP binding / nucleus / metal ion binding / cytosol / cytoplasm 類似検索 - 分子機能 | ||||||||||||||||||
| 生物種 |  | ||||||||||||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.6 Å | ||||||||||||||||||
 データ登録者 データ登録者 | Schilbach S / Aibara S | ||||||||||||||||||
| 資金援助 |  ドイツ, 5件 ドイツ, 5件
| ||||||||||||||||||
 引用 引用 |  ジャーナル: Cell / 年: 2021 ジャーナル: Cell / 年: 2021タイトル: Structure of RNA polymerase II pre-initiation complex at 2.9 Å defines initial DNA opening. 著者: Sandra Schilbach / Shintaro Aibara / Christian Dienemann / Frauke Grabbe / Patrick Cramer /  要旨: Transcription initiation requires assembly of the RNA polymerase II (Pol II) pre-initiation complex (PIC) and opening of promoter DNA. Here, we present the long-sought high-resolution structure of ...Transcription initiation requires assembly of the RNA polymerase II (Pol II) pre-initiation complex (PIC) and opening of promoter DNA. Here, we present the long-sought high-resolution structure of the yeast PIC and define the mechanism of initial DNA opening. We trap the PIC in an intermediate state that contains half a turn of open DNA located 30-35 base pairs downstream of the TATA box. The initially opened DNA region is flanked and stabilized by the polymerase "clamp head loop" and the TFIIF "charged region" that both contribute to promoter-initiated transcription. TFIIE facilitates initiation by buttressing the clamp head loop and by regulating the TFIIH translocase. The initial DNA bubble is then extended in the upstream direction, leading to the open promoter complex and enabling start-site scanning and RNA synthesis. This unique mechanism of DNA opening may permit more intricate regulation than in the Pol I and Pol III systems. | ||||||||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
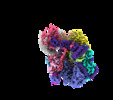
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_12721.map.gz emd_12721.map.gz | 187.5 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-12721-v30.xml emd-12721-v30.xml emd-12721.xml emd-12721.xml | 77 KB 77 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_12721_fsc.xml emd_12721_fsc.xml | 13.5 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_12721.png emd_12721.png | 83.9 KB | ||
| マスクデータ |  emd_12721_msk_1.map emd_12721_msk_1.map | 209.3 MB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-12721.cif.gz emd-12721.cif.gz | 13.8 KB | ||
| その他 |  emd_12721_additional_1.map.gz emd_12721_additional_1.map.gz emd_12721_additional_10.map.gz emd_12721_additional_10.map.gz emd_12721_additional_11.map.gz emd_12721_additional_11.map.gz emd_12721_additional_12.map.gz emd_12721_additional_12.map.gz emd_12721_additional_13.map.gz emd_12721_additional_13.map.gz emd_12721_additional_14.map.gz emd_12721_additional_14.map.gz emd_12721_additional_15.map.gz emd_12721_additional_15.map.gz emd_12721_additional_2.map.gz emd_12721_additional_2.map.gz emd_12721_additional_3.map.gz emd_12721_additional_3.map.gz emd_12721_additional_4.map.gz emd_12721_additional_4.map.gz emd_12721_additional_5.map.gz emd_12721_additional_5.map.gz emd_12721_additional_6.map.gz emd_12721_additional_6.map.gz emd_12721_additional_7.map.gz emd_12721_additional_7.map.gz emd_12721_additional_8.map.gz emd_12721_additional_8.map.gz emd_12721_additional_9.map.gz emd_12721_additional_9.map.gz emd_12721_half_map_1.map.gz emd_12721_half_map_1.map.gz emd_12721_half_map_2.map.gz emd_12721_half_map_2.map.gz | 121.2 MB 95.5 MB 95.8 MB 95.6 MB 95.9 MB 94.3 MB 94.2 MB 96 MB 94.5 MB 94.4 MB 96 MB 95.9 MB 95.9 MB 95.7 MB 95.5 MB 103.1 MB 102.8 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12721 http://ftp.pdbj.org/pub/emdb/structures/EMD-12721 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12721 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12721 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_12721_validation.pdf.gz emd_12721_validation.pdf.gz | 740.6 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_12721_full_validation.pdf.gz emd_12721_full_validation.pdf.gz | 740.2 KB | 表示 | |
| XML形式データ |  emd_12721_validation.xml.gz emd_12721_validation.xml.gz | 20.7 KB | 表示 | |
| CIF形式データ |  emd_12721_validation.cif.gz emd_12721_validation.cif.gz | 26.6 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12721 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12721 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12721 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12721 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_12721.map.gz / 形式: CCP4 / 大きさ: 209.3 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_12721.map.gz / 形式: CCP4 / 大きさ: 209.3 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
+マスク #1
+追加マップ: Locally filtered map (non-composite) of a refinement of...
+追加マップ: Half-map 2 of focused refinement encompassing the Tfb2...
+追加マップ: Half-map 2 of focused refinement encompassing the Ssl2/Tfb2/Tfb5...
+追加マップ: Half-map 1 of focused refinement encompassing the Tfb2...
+追加マップ: Half-map 1 of focused refinement encompassing the Ssl2/Tfb2/Tfb5...
+追加マップ: Half-map 1 of focused refinement encompassing the Tfb3-RING-finger/TFIIE/Pol...
+追加マップ: Half-map 2 of focused refinement encompassing the Tfb3-RING-finger/TFIIE/Pol...
+追加マップ: Half-map 1 of focused refinement encompassing the Pol...
+追加マップ: Half-map 1 of focused refinement encompassing the Pol...
+追加マップ: Half-map 2 of focused refinement encompassing the Pol...
+追加マップ: Half-map 2 of focused refinement encompassing the Pol...
+追加マップ: Half-map 1 of focused refinement encompassing the Rad3/Ssl1...
+追加マップ: Half-map 2 of focused refinement encompassing the Rad3/Ssl1...
+追加マップ: Half-map 1 of focused refinement encompassing the Ssl1/Tfb4-eZnF/Tfb1-3-helix-bundle...
+追加マップ: Half-map 2 of focused refinement encompassing the Ssl1/Tfb4-eZnF/Tfb1-3-helix-bundle...
+ハーフマップ: #2
+ハーフマップ: #1
- 試料の構成要素
試料の構成要素
+全体 : Yeast TFIIH in the contracted state within the pre-initiation complex
+超分子 #1: Yeast TFIIH in the contracted state within the pre-initiation complex
+分子 #1: General transcription and DNA repair factor IIH helicase subunit XPD
+分子 #2: General transcription and DNA repair factor IIH subunit TFB1
+分子 #3: General transcription and DNA repair factor IIH subunit TFB2
+分子 #4: RNA polymerase II transcription factor B subunit 3
+分子 #5: General transcription and DNA repair factor IIH subunit TFB4
+分子 #6: General transcription and DNA repair factor IIH subunit TFB5
+分子 #7: General transcription and DNA repair factor IIH subunit SSL1
+分子 #8: General transcription and DNA repair factor IIH helicase subunit XPB
+分子 #9: DNA-directed RNA polymerase II subunit RPB1
+分子 #10: DNA-directed RNA polymerase II subunit RPB2
+分子 #11: DNA-directed RNA polymerase II subunit RPB4
+分子 #12: DNA-directed RNA polymerases I, II, and III subunit RPABC2
+分子 #13: DNA-directed RNA polymerase II subunit RPB7
+分子 #16: Transcription initiation factor IIE subunit alpha
+分子 #17: Transcription initiation factor IIE subunit beta
+分子 #14: Non-template DNA
+分子 #15: Template DNA
+分子 #18: IRON/SULFUR CLUSTER
+分子 #19: ZINC ION
+分子 #20: BERYLLIUM TRIFLUORIDE ION
+分子 #21: MAGNESIUM ION
+分子 #22: ADENOSINE-5'-DIPHOSPHATE
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.6 |
|---|---|
| グリッド | モデル: Quantifoil R3.5/1 / 材質: COPPER / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: CONTINUOUS |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 277 K / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 特殊光学系 | エネルギーフィルター - 名称: GIF Quantum LS / エネルギーフィルター - スリット幅: 20 eV |
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: COUNTING / 実像数: 29670 / 平均露光時間: 9.0 sec. / 平均電子線量: 43.6 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 4.0 µm / 最小 デフォーカス(公称値): 0.5 µm / 倍率(公称値): 130000 |
| 試料ステージ | ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー










































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)






































































































































 Trichoplusia ni (イラクサキンウワバ)
Trichoplusia ni (イラクサキンウワバ)