[English] 日本語
 Yorodumi
Yorodumi- EMDB-10944: Head segment of the S.cerevisiae condensin holocomplex in presenc... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10944 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Head segment of the S.cerevisiae condensin holocomplex in presence of ATP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Condensin chromosome condensation SMC protein / CELL CYCLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of meiotic DNA double-strand break formation / Condensation of Prometaphase Chromosomes / meiotic chromosome condensation / tRNA gene clustering / meiotic chromosome separation / condensin complex / DNA secondary structure binding / rDNA chromatin condensation / synaptonemal complex assembly / mitotic chromosome condensation ...negative regulation of meiotic DNA double-strand break formation / Condensation of Prometaphase Chromosomes / meiotic chromosome condensation / tRNA gene clustering / meiotic chromosome separation / condensin complex / DNA secondary structure binding / rDNA chromatin condensation / synaptonemal complex assembly / mitotic chromosome condensation / chromosome condensation / minor groove of adenine-thymine-rich DNA binding / mitotic sister chromatid segregation / condensed chromosome / double-stranded DNA binding / cell division / chromatin binding / chromatin / ATP hydrolysis activity / mitochondrion / ATP binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
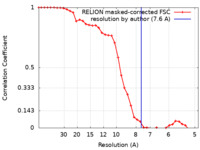
| Method | single particle reconstruction / cryo EM / Resolution: 7.6 Å | |||||||||
 Authors Authors | Merkel F / Haering CH | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2020 Journal: Nat Struct Mol Biol / Year: 2020Title: Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism. Authors: Byung-Gil Lee / Fabian Merkel / Matteo Allegretti / Markus Hassler / Christopher Cawood / Léa Lecomte / Francis J O'Reilly / Ludwig R Sinn / Pilar Gutierrez-Escribano / Marc Kschonsak / Sol ...Authors: Byung-Gil Lee / Fabian Merkel / Matteo Allegretti / Markus Hassler / Christopher Cawood / Léa Lecomte / Francis J O'Reilly / Ludwig R Sinn / Pilar Gutierrez-Escribano / Marc Kschonsak / Sol Bravo / Takanori Nakane / Juri Rappsilber / Luis Aragon / Martin Beck / Jan Löwe / Christian H Haering /    Abstract: Complexes containing a pair of structural maintenance of chromosomes (SMC) family proteins are fundamental for the three-dimensional (3D) organization of genomes in all domains of life. The ...Complexes containing a pair of structural maintenance of chromosomes (SMC) family proteins are fundamental for the three-dimensional (3D) organization of genomes in all domains of life. The eukaryotic SMC complexes cohesin and condensin are thought to fold interphase and mitotic chromosomes, respectively, into large loop domains, although the underlying molecular mechanisms have remained unknown. We used cryo-EM to investigate the nucleotide-driven reaction cycle of condensin from the budding yeast Saccharomyces cerevisiae. Our structures of the five-subunit condensin holo complex at different functional stages suggest that ATP binding induces the transition of the SMC coiled coils from a folded-rod conformation into a more open architecture. ATP binding simultaneously triggers the exchange of the two HEAT-repeat subunits bound to the SMC ATPase head domains. We propose that these steps result in the interconversion of DNA-binding sites in the catalytic core of condensin, forming the basis of the DNA translocation and loop-extrusion activities. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10944.map.gz emd_10944.map.gz | 576.5 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10944-v30.xml emd-10944-v30.xml emd-10944.xml emd-10944.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10944_fsc.xml emd_10944_fsc.xml | 3.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_10944.png emd_10944.png | 80.2 KB | ||
| Masks |  emd_10944_msk_1.map emd_10944_msk_1.map | 3.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10944.cif.gz emd-10944.cif.gz | 8.5 KB | ||
| Others |  emd_10944_half_map_1.map.gz emd_10944_half_map_1.map.gz emd_10944_half_map_2.map.gz emd_10944_half_map_2.map.gz | 2.8 MB 2.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10944 http://ftp.pdbj.org/pub/emdb/structures/EMD-10944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10944 | HTTPS FTP |
-Validation report
| Summary document |  emd_10944_validation.pdf.gz emd_10944_validation.pdf.gz | 648.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10944_full_validation.pdf.gz emd_10944_full_validation.pdf.gz | 647.7 KB | Display | |
| Data in XML |  emd_10944_validation.xml.gz emd_10944_validation.xml.gz | 8.9 KB | Display | |
| Data in CIF |  emd_10944_validation.cif.gz emd_10944_validation.cif.gz | 11.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10944 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10944 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10944 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10944 | HTTPS FTP |
-Related structure data
| Related structure data |  6yvdMC  6yvuC  6yvvC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10944.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10944.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.448 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10944_msk_1.map emd_10944_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_10944_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_10944_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : condensin
| Entire | Name: condensin |
|---|---|
| Components |
|
-Supramolecule #1: condensin
| Supramolecule | Name: condensin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Engaged form in present of nucleotide |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 650 KDa |
-Macromolecule #1: Condensin complex subunit 2
| Macromolecule | Name: Condensin complex subunit 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 92.730164 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTTQLRYENN DDDERVEYNL FTNRSTMMAN FEEWIKMATD NKINSRNSWN FALIDYFYDL DVLKDGENNI NFQKASATLD GCIKIYSSR VDSVTTETGK LLSGLAQRKT NGASNGDDSN GGNGEGLGGD SDEANIEIDP LTGMPISNDP DVNNTRRRVY N RVLETTLV ...String: MTTQLRYENN DDDERVEYNL FTNRSTMMAN FEEWIKMATD NKINSRNSWN FALIDYFYDL DVLKDGENNI NFQKASATLD GCIKIYSSR VDSVTTETGK LLSGLAQRKT NGASNGDDSN GGNGEGLGGD SDEANIEIDP LTGMPISNDP DVNNTRRRVY N RVLETTLV EFETIKMKEL DQELIIDPLF KKALVDFDEG GAKSLLLNTL NIDNTARVIF DASIKDTQNV GQGKLQRKEE EL IERDSLV DDENEPSQSL ISTRNDSTVN DSVISAPSME DEILSLGMDF IKFDQIAVCE ISGSIEQLRN VVEDINQAKD FIE NVNNRF DNFLTEEELQ AAVPDNAEDD SDGFDMGMQQ ELCYPDENHD NTSHDEQDDD NVNSTTGSIF EKDLMAYFDE NLNR NWRGR EHWKVRNFKK ANLVNKESDL LEETRTTIGD TTDKNTTDDK SMDTKKKHKQ KKVLEIDFFK TDDSFEDKVF ASKGR TKID MPIKNRKNDT HYLLPDDFHF STDRITRLFI KPGQKMSLFS HRKHTRGDVS SGLFEKSTVS ANHSNNDIPT IADEHF WAD NYERKEQEEK EKEQSKEVGD VVGGALDNPF EDDMDGVDFN QAFEGTDDNE EASVKLDLQD DEDHKFPIRE NKVTYSR VS KKVDVRRLKK NVWRSINNLI QEHDSRKNRE QSSNDSETHT EDESTKELKF SDIIQGISKM YSDDTLKDIS TSFCFICL L HLANEHGLQI THTENYNDLI VNYEDLATTQ AASLVGGGHH HHHHGGHHHH HHGGRIFYPY DVPDYAGYPY DVPDYAGSY PYDVPDYAAG H UniProtKB: Condensin complex subunit 2 |
-Macromolecule #2: Condensin complex subunit 3
| Macromolecule | Name: Condensin complex subunit 3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 117.981 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQDPDGIDIN TKIFNSVAEV FQKAQGSYAG HRKHIAVLKK IQSKAVEQGY EDAFNFWFDK LVTKILPLKK NEIIGDRIVK LVAAFIASL ERELILAKKQ NYKLTNDEEG IFSRFVDQFI RHVLRGVESP DKNVRFRVLQ LLAVIMDNIG EIDESLFNLL I LSLNKRIY ...String: MQDPDGIDIN TKIFNSVAEV FQKAQGSYAG HRKHIAVLKK IQSKAVEQGY EDAFNFWFDK LVTKILPLKK NEIIGDRIVK LVAAFIASL ERELILAKKQ NYKLTNDEEG IFSRFVDQFI RHVLRGVESP DKNVRFRVLQ LLAVIMDNIG EIDESLFNLL I LSLNKRIY DREPTVRIQA VFCLTKFQDE EQTEHLTELS DNEENFEATR TLVASIQNDP SAEVRRAAML NLINDNNTRP YI LERARDV NIVNRRLVYS RILKSMGRKC FDDIEPHIFD QLIEWGLEDR ELSVRNACKR LIAHDWLNAL DGDLIELLEK LDV SRSSVC VKAIEALFQS RPDILSKIKF PESIWKDFTV EIAFLFRAIY LYCLDNNITE MLEENFPEAS KLSEHLNHYI LLRY HHNDI SNDSQSHFDY NTLEFIIEQL SIAAERYDYS DEVGRRSMLT VVRNMLALTT LSEPLIKIGI RVMKSLSINE KDFVT MAIE IINDIRDDDI EKQEQEEKIK SKKINRRNET SVDEEDENGT HNDEVNEDEE DDNISSFHSA VENLVQGNGN VSESDI INN LPPEKEASSA TIVLCLTRSS YMLELVNTPL TENILIASLM DTLITPAVRN TAPNIRELGV KNLGLCCLLD VKLAIDN MY ILGMCVSKGN ASLKYIALQV IVDIFSVHGN TVVDGEGKVD SISLHKIFYK VLKNNGLPEC QVIAAEGLCK LFLADVFT D DDLFETLVLS YFSPINSSNE ALVQAFAFCI PVYCFSHPAH QQRMSRTAAD ILLRLCVLWD DLQSSVIPEV DREAMLKPN IIFQQLLFWT DPRNLVNQTG STKKDTVQLT FLIDVLKIYA QIEKKEIKKM IITNINAIFL SSEQDYSTLK ELLEYSDDIA ENDNLDNVS KNALDKLRNN LNSLIEEINE RSETQTKDEN NTANDQYSSI LGNSFNKSSN DTIEHAADIT DGNNTELTKT T VNISAVDN TTEQSNSRKR TRSEAEQIDT SKNLENMSIQ DTSTVAKNVS FVLPDEKSDA MSIDEEDKDS ESFSEVC UniProtKB: Condensin complex subunit 3 |
-Macromolecule #3: Structural maintenance of chromosomes protein 2
| Macromolecule | Name: Structural maintenance of chromosomes protein 2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 134.125875 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKVEELIIDG FKSYATRTVI TDWDPQFNAI TGLNGSGKSN ILDAICFVLG IASMSTVRAS SLQDLIYKRG QAGVTKASVT IVFDNTDKS NSPIGFTNSP QISVTRQVVL GGTSKYLING HRAPQQSVLQ LFQSVQLNIN NPNFLIMQGK ITKVLNMKPS E ILSLIEEA ...String: MKVEELIIDG FKSYATRTVI TDWDPQFNAI TGLNGSGKSN ILDAICFVLG IASMSTVRAS SLQDLIYKRG QAGVTKASVT IVFDNTDKS NSPIGFTNSP QISVTRQVVL GGTSKYLING HRAPQQSVLQ LFQSVQLNIN NPNFLIMQGK ITKVLNMKPS E ILSLIEEA AGTKMFEDRR EKAERTMSKK ETKLQENRTL LTEEIEPKLE KLRNEKRMFL EFQSTQTDLE KTERIVVSYE YY NIKHKHT SIRETLENGE TRMKMLNEFV KKTSEEIDSL NEDVEEIKLQ KEKELHKEGT ISKLENKENG LLNEISRLKT SLS IKVENL NDTTEKSKAL ESEIASSSAK LIEKKSAYAN TEKDYKMVQE QLSKQRDLYK RKEELVSTLT TGISSTGAAD GGYN AQLAK AKTELNEVSL AIKKSSMKME LLKKELLTIE PKLKEATKDN ELNVKHVKQC QETCDKLRAR LVEYGFDPSR IKDLK QRED KLKSHYYQTC KNSEYLKRRV TNLEFNYTKP YPNFEASFVH GVVGQLFQID NDNIRYATAL QTCAGGRLFN VVVQDS QTA TQLLERGRLR KRVTIIPLDK IYTRPISSQV LDLAKKIAPG KVELAINLIR FDESITKAME FIFGNSLICE DPETAKK IT FHPKIRARSI TLQGDVYDPE GTLSGGSRNT SESLLVDIQK YNQIQKQIET IQADLNHVTE ELQTQYATSQ KTKTIQSD L NLSLHKLDLA KRNLDANPSS QIIARNEEIL RDIGECENEI KTKQMSLKKC QEEVSTIEKD MKEYDSDKGS KLNELKKEL KLLAKELEEQ ESESERKYDL FQNLELETEQ LSSELDSNKT LLHNHLKSIE SLKLENSDLE GKIRGVEDDL VTVQTELNEE KKRLMDIDD ELNELETLIK KKQDEKKSSE LELQKLVHDL NKYKSNTNNM EKIIEDLRQK HEFLEDFDLV RNIVKQNEGI D LDTYRERS KQLNEKFQEL RKKVNPNIMN MIENVEKKEA ALKTMIKTIE KDKMKIQETI SKLNEYKRET LVKTWEKVTL DF GNIFADL LPNSFAKLVP CEGKDVTQGL EVKVKLGNIW KESLIELSGG QRSLIALSLI MALLQFRPAP MYILDEVDAA LDL SHTQNI GHLIKTRFKG SQFIVVSLKE GMFANANRVF RTRFQDGTSV VSIM UniProtKB: Structural maintenance of chromosomes protein 2 |
-Macromolecule #4: Structural maintenance of chromosomes protein 4
| Macromolecule | Name: Structural maintenance of chromosomes protein 4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 168.192625 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSDSPLSKRQ KRKAAQEPEL SLDQGDAEEE SQVENRVNLS ENTPEPDLPA LEASYSKSYT PRKLVLSSGE NRYAFSQPTN STTTSLHVP NLQPPKTSSR GRDHKSYSQS PPRSPGRSPT RRLELLQLSP VKNSRVELQK IYDSHQSSSK QQSRLFINEL V LENFKSYA ...String: MSDSPLSKRQ KRKAAQEPEL SLDQGDAEEE SQVENRVNLS ENTPEPDLPA LEASYSKSYT PRKLVLSSGE NRYAFSQPTN STTTSLHVP NLQPPKTSSR GRDHKSYSQS PPRSPGRSPT RRLELLQLSP VKNSRVELQK IYDSHQSSSK QQSRLFINEL V LENFKSYA GKQVVGPFHT SFSAVVGPNG SGKSNVIDSM LFVFGFRANK MRQDRLSDLI HKSEAFPSLQ SCSVAVHFQY VI DESSGTS RIDEEKPGLI ITRKAFKNNS SKYYINEKES SYTEVTKLLK NEGIDLDHKR FLILQGEVEN IAQMKPKAEK ESD DGLLEY LEDIIGTANY KPLIEERMGQ IENLNEVCLE KENRFEIVDR EKNSLESGKE TALEFLEKEK QLTLLRSKLF QFKL LQSNS KLASTLEKIS SSNKDLEDER MKFQESLKKV DEIKAQRKEI KDRISSCSSK EKTLVLERRE LEGTRVSLEE RTKNL VSKM EKAEKTLKST KHSISEAENM LEELRGQQTE HETEIKDLTQ LLEKERSILD DIKLSLKDKT KDISAEIIRH EKELEP WDL QLQEKESQIQ LAESELSLLE ETQAKLKKNV ETLEEKILAK KTHKQELQDL ILDLKKKLNS LKDERSQGEK NFTSAHL KL KEMQKVLNAH RQRAMEARSS LSKAQNKSKV LTALSRLQKS GRINGFHGRL GDLGAIDDSF DIAISTACPR LDDVVVDT V ECAQHCIDYL RKNKLGYARF ILLDRLRQFN LQPISTPENV PRLFDLVKPK NPKFSNAFYS VLRDTLVAQN LKQANNVAY GKKRFRVVTV DGKLIDISGT MSGGGNHVAK GLMKLGTNQS DKVDDYTPEE VDKIERELSE RENNFRVASD TVHEMEEELK KLRDHEPDL ESQISKAEME ADSLASELTL AEQQVKEAEM AYVKAVSDKA QLNVVMKNLE RLRGEYNDLQ SETKTKKEKI K GLQDEIMK IGGIKLQMQN SKVESVCQKL DILVAKLKKV KSASKKSGGD VVKFQKLLQN SERDVELSSN ELKVIEEQLK HT KLALAEN DTNMTETLNL KVELKEQSEQ LKEQMEDMEE SINEFKSIEI EMKNKLEKLN SLLTYIKSEI TQQEKGLNEL SIR DVTHTL GMLDDNKMDS VKEDVKNNQE LDQEYRSCET QDESEIKDDE TSCDNYHPMN VDETSDEVSR GIPRLSEDEL RELD VELIE SKINELSYYV EETNVDIGVL EEYARRLAEF KRRKLDLNNA VQKRDEVKEQ LGILKKKRFD EFMAGFNIIS MTLKE MYQM ITMGGNAELE LVDSLDPFSE GVTFSVMPPK KSWRNITNLS GGEKTLSSLA LVFALHKYKP TPLYVMDEID AALDFR NVS IVANYIKERT KNAQFIVISL RNNMFELAQQ LVGVYKRDNR TKSTTIKNID ILNRTRIPGL INGATGWSHP QFEKAGG GS GGGSGGGSWS HPQFEKGGGS GGGSGGGSWS HPQFEK UniProtKB: Structural maintenance of chromosomes protein 4 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 200 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)