[English] 日本語
 Yorodumi
Yorodumi- EMDB-0441: Conformational switches control early maturation of the eukaryoti... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0441 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Conformational switches control early maturation of the eukaryotic small ribosomal subunit | |||||||||
 Map data Map data | masked, sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribosome assembly / RIBOSOME | |||||||||
| Function / homology |  Function and homology information Function and homology informationtRNA re-export from nucleus / RNA fragment catabolic process / rRNA 2'-O-methylation / t-UTP complex / Mpp10 complex / snoRNA guided rRNA 2'-O-methylation / Pwp2p-containing subcomplex of 90S preribosome / rRNA modification / histone H2AQ104 methyltransferase activity / positive regulation of RNA binding ...tRNA re-export from nucleus / RNA fragment catabolic process / rRNA 2'-O-methylation / t-UTP complex / Mpp10 complex / snoRNA guided rRNA 2'-O-methylation / Pwp2p-containing subcomplex of 90S preribosome / rRNA modification / histone H2AQ104 methyltransferase activity / positive regulation of RNA binding / snRNA binding / endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / septum digestion after cytokinesis / box C/D sno(s)RNA 3'-end processing / rDNA heterochromatin / tRNA export from nucleus / rRNA methyltransferase activity / endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / regulation of transcription by RNA polymerase I / positive regulation of rRNA processing / U4/U6 snRNP / O-methyltransferase activity / SUMOylation of RNA binding proteins / protein localization to nucleolus / U4 snRNA binding / sno(s)RNA-containing ribonucleoprotein complex / box C/D methylation guide snoRNP complex / rRNA methylation / U4 snRNP / U3 snoRNA binding / preribosome, small subunit precursor / establishment of cell polarity / snoRNA binding / precatalytic spliceosome / positive regulation of transcription by RNA polymerase I / Major pathway of rRNA processing in the nucleolus and cytosol / spliceosomal complex assembly / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / U4/U6 x U5 tri-snRNP complex / RNA endonuclease activity / 90S preribosome / Transferases; Transferring one-carbon groups; Methyltransferases / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of LSU-rRNA / maturation of SSU-rRNA / small-subunit processome / spliceosomal complex / mRNA splicing, via spliceosome / rRNA processing / peroxisome / ribosomal small subunit biogenesis / ribosomal small subunit assembly / small ribosomal subunit / cytosolic large ribosomal subunit / tRNA binding / rRNA binding / structural constituent of ribosome / mRNA binding / nucleolus / mitochondrion / RNA binding / nucleoplasm / identical protein binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
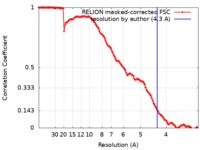
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Hunziker M / Barandun J | |||||||||
| Funding support |  United States, United States,  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: Conformational switches control early maturation of the eukaryotic small ribosomal subunit. Authors: Mirjam Hunziker / Jonas Barandun / Olga Buzovetsky / Caitlin Steckler / Henrik Molina / Sebastian Klinge /  Abstract: Eukaryotic ribosome biogenesis is initiated with the transcription of pre-ribosomal RNA at the 5' external transcribed spacer, which directs the early association of assembly factors but is absent ...Eukaryotic ribosome biogenesis is initiated with the transcription of pre-ribosomal RNA at the 5' external transcribed spacer, which directs the early association of assembly factors but is absent from the mature ribosome. The subsequent co-transcriptional association of ribosome assembly factors with pre-ribosomal RNA results in the formation of the small subunit processome. Here we show that stable rRNA domains of the small ribosomal subunit can independently recruit their own biogenesis factors in vivo. The final assembly and compaction of the small subunit processome requires the presence of the 5' external transcribed spacer RNA and all ribosomal RNA domains. Additionally, our cryo-electron microscopy structure of the earliest nucleolar pre-ribosomal assembly - the 5' external transcribed spacer ribonucleoprotein - provides a mechanism for how conformational changes in multi-protein complexes can be employed to regulate the accessibility of binding sites and therefore define the chronology of maturation events during early stages of ribosome assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0441.map.gz emd_0441.map.gz | 10.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0441-v30.xml emd-0441-v30.xml emd-0441.xml emd-0441.xml | 57 KB 57 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0441_fsc.xml emd_0441_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_0441.png emd_0441.png | 179.5 KB | ||
| Filedesc metadata |  emd-0441.cif.gz emd-0441.cif.gz | 15.9 KB | ||
| Others |  emd_0441_additional.map.gz emd_0441_additional.map.gz | 140.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0441 http://ftp.pdbj.org/pub/emdb/structures/EMD-0441 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0441 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0441 | HTTPS FTP |
-Validation report
| Summary document |  emd_0441_validation.pdf.gz emd_0441_validation.pdf.gz | 395.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0441_full_validation.pdf.gz emd_0441_full_validation.pdf.gz | 395 KB | Display | |
| Data in XML |  emd_0441_validation.xml.gz emd_0441_validation.xml.gz | 12.9 KB | Display | |
| Data in CIF |  emd_0441_validation.cif.gz emd_0441_validation.cif.gz | 17.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0441 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0441 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0441 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0441 | HTTPS FTP |
-Related structure data
| Related structure data |  6nd4MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0441.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0441.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | masked, sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.6 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: unsharpened map
| File | emd_0441_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : 5' ETS particle
+Supramolecule #1: 5' ETS particle
+Macromolecule #1: 5'ETS rRNA
+Macromolecule #2: 18S rRNA 5' domain start
+Macromolecule #3: U3 snoRNA
+Macromolecule #4: Utp17
+Macromolecule #5: Utp8
+Macromolecule #6: Utp15
+Macromolecule #7: Utp9
+Macromolecule #8: Utp5
+Macromolecule #9: Utp10
+Macromolecule #10: Utp4
+Macromolecule #11: Utp1
+Macromolecule #12: Utp6
+Macromolecule #13: Utp12
+Macromolecule #14: Utp13
+Macromolecule #15: Utp18
+Macromolecule #16: Utp21
+Macromolecule #17: Sof1
+Macromolecule #18: Utp7
+Macromolecule #19: Imp3
+Macromolecule #20: Mpp10
+Macromolecule #21: Bud21
+Macromolecule #22: Nop56
+Macromolecule #23: Nop58
+Macromolecule #24: Nop1.1
+Macromolecule #25: Snu13
+Macromolecule #26: Rrp9
+Macromolecule #27: Utp24
+Macromolecule #28: Unidentified fragment
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.7 Component:
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Support film - #0 - Film type ID: 1 / Support film - #0 - Material: CARBON / Support film - #0 - topology: LACEY / Support film - #1 - Film type ID: 2 / Support film - #1 - Material: CARBON / Support film - #1 - topology: CONTINUOUS / Support film - #1 - Film thickness: 2 | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-32 / Number grids imaged: 1 / Number real images: 2750 / Average exposure time: 0.25 sec. / Average electron dose: 31.25 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z
Z Y
Y X
X