[English] 日本語
 Yorodumi
Yorodumi- EMDB-8981: Cryo-EM reconstruction of domain-swapped, glycan-reactive, neutra... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8981 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reconstruction of domain-swapped, glycan-reactive, neutralizing antibody 2G12 bound to HIV-1 Env BG505 DS-SOSIP, which was also bound to CD4-binding site antibody VRC03 | |||||||||
 Map data Map data | Refined map from cryoSparc postprocessed in Relion using map fro cryoSparc | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 Env / complex / neutralizing / VIRAL PROTEIN / domain-swapped antibody | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
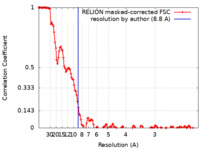
| Method | single particle reconstruction / cryo EM / Resolution: 8.8 Å | |||||||||
 Authors Authors | Acharya P / Kwong PD | |||||||||
 Citation Citation |  Journal: Structure / Year: 2019 Journal: Structure / Year: 2019Title: Structural Survey of Broadly Neutralizing Antibodies Targeting the HIV-1 Env Trimer Delineates Epitope Categories and Characteristics of Recognition. Authors: Gwo-Yu Chuang / Jing Zhou / Priyamvada Acharya / Reda Rawi / Chen-Hsiang Shen / Zizhang Sheng / Baoshan Zhang / Tongqing Zhou / Robert T Bailer / Venkata P Dandey / Nicole A Doria-Rose / ...Authors: Gwo-Yu Chuang / Jing Zhou / Priyamvada Acharya / Reda Rawi / Chen-Hsiang Shen / Zizhang Sheng / Baoshan Zhang / Tongqing Zhou / Robert T Bailer / Venkata P Dandey / Nicole A Doria-Rose / Mark K Louder / Krisha McKee / John R Mascola / Lawrence Shapiro / Peter D Kwong /  Abstract: Over the past decade, structures have been determined for broadly neutralizing antibodies that recognize all major exposed surfaces of the prefusion-closed HIV-1-envelope (Env) trimer. To understand ...Over the past decade, structures have been determined for broadly neutralizing antibodies that recognize all major exposed surfaces of the prefusion-closed HIV-1-envelope (Env) trimer. To understand this recognition and its implications, we analyzed 206 antibody-HIV-1 Env structures from the Protein Data Bank with resolution suitable to define interaction chemistries and measured antibody neutralization on a 208-strain panel. Those with >25% breadth segregated into almost two dozen classes based on ontogeny and recognition and into six epitope categories based on recognized Env residues. For paratope, the number of protruding loops and level of somatic hypermutation were significantly higher for broad HIV-1 neutralizing antibodies than for a comparison set of non-HIV-1 antibodies (p < 0.0001). For epitope, the number of independent sequence segments was higher (p < 0.0001), as well as the glycan component surface area (p = 0.0005). The unusual characteristics of epitope and paratope delineated here are likely to reflect respectively virus-immune evasion and antibody-recognition solutions that allow effective neutralization of HIV-1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8981.map.gz emd_8981.map.gz | 11.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8981-v30.xml emd-8981-v30.xml emd-8981.xml emd-8981.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8981_fsc.xml emd_8981_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_8981.png emd_8981.png | 94.7 KB | ||
| Filedesc metadata |  emd-8981.cif.gz emd-8981.cif.gz | 7.2 KB | ||
| Others |  emd_8981_additional.map.gz emd_8981_additional.map.gz emd_8981_additional_1.map.gz emd_8981_additional_1.map.gz | 105.2 MB 105.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8981 http://ftp.pdbj.org/pub/emdb/structures/EMD-8981 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8981 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8981 | HTTPS FTP |
-Related structure data
| Related structure data |  6e5pMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8981.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8981.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refined map from cryoSparc postprocessed in Relion using map fro cryoSparc | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0961 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Unsharpened map from cryoSparc
| File | emd_8981_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map from cryoSparc | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened map from cryoSparc
| File | emd_8981_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map from cryoSparc | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex HIV-1 Env BG505 DS-SOSIP with Fab fragments of an...
| Entire | Name: Ternary complex HIV-1 Env BG505 DS-SOSIP with Fab fragments of antibodies 2G12 and VRC03. |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex HIV-1 Env BG505 DS-SOSIP with Fab fragments of an...
| Supramolecule | Name: Ternary complex HIV-1 Env BG505 DS-SOSIP with Fab fragments of antibodies 2G12 and VRC03. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: 2G12 Light chain
| Macromolecule | Name: 2G12 Light chain / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.130762 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VVMTQSPSTL SASVGDTITI TCRASQSIET WLAWYQQKPG KAPKLLIYKA STLKTGVPSR FSGSGSGTEF TLTISGLQFD DFATYHCQH YAGYSATFGQ GTRVEIKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ E SVTEQDSK ...String: VVMTQSPSTL SASVGDTITI TCRASQSIET WLAWYQQKPG KAPKLLIYKA STLKTGVPSR FSGSGSGTEF TLTISGLQFD DFATYHCQH YAGYSATFGQ GTRVEIKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ E SVTEQDSK DSTYSLSSTL TLSKADYEKH KVYACEVTHQ GLSSPVTKSF NRGE |
-Macromolecule #2: 2G12 heavy chain
| Macromolecule | Name: 2G12 heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.932869 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LVKAGGSLIL SCGVSNFRIS AHTMNWVRRV PGGGLEWVAS ISTSSTYRDY ADAVKGRFTV SRDDLEDFVY LQMHKMRVE DTAIYYCARK GSDRLSDNDP FDAWGPGTVV TVSPASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA ...String: EVQLVESGGG LVKAGGSLIL SCGVSNFRIS AHTMNWVRRV PGGGLEWVAS ISTSSTYRDY ADAVKGRFTV SRDDLEDFVY LQMHKMRVE DTAIYYCARK GSDRLSDNDP FDAWGPGTVV TVSPASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KKVEPKS |
-Macromolecule #3: Envelope glycoprotein gp120
| Macromolecule | Name: Envelope glycoprotein gp120 / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 53.086102 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT ...String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT SACTQACPKV SFEPIPIHYC APAGFAILKC KDKKFNGTGP CPSVSTVQCT HGIKPVVSTQ LLLNGSLAEE EV MIRSENI TNNAKNILVQ FNTPVQINCT RPNNNTRKSI RIGPGQAFYA TGDIIGDIRQ AHCNVSKATW NETLGKVVKQ LRK HFGNNT IIRFANSSGG DLEVTTHSFN CGGEFFYCNT SGLFNSTWIS NTSVQGSNST GSNDSITLPC RIKQIINMWQ RIGQ CMYAP PIQGVIRCVS NITGLILTRD GGSTNSTTET FRPGGGDMRD NWRSELYKYK VVKIEPLGVA PTRCKRRVV UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #4: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 19.102811 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEA QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALDGSAPTK A KRRVVQRE KR UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #5: VRC03 heavy chain
| Macromolecule | Name: VRC03 heavy chain / type: protein_or_peptide / ID: 5 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.699861 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAV IKTPGSSVKI SCRASGYNFR DYSIHWVRLI PDKGFEWIGW IKPLWGAVSY ARQLQGRVSM TRQLSQDPDD PDWGVAYME FSGLTPADTA EYFCVRRGSC DYCGDFPWQY WCQGTVVVVS SASTKGPSVF PLAPSSGGTA ALGCLVKDYF P EPVTVSWN ...String: QVQLVQSGAV IKTPGSSVKI SCRASGYNFR DYSIHWVRLI PDKGFEWIGW IKPLWGAVSY ARQLQGRVSM TRQLSQDPDD PDWGVAYME FSGLTPADTA EYFCVRRGSC DYCGDFPWQY WCQGTVVVVS SASTKGPSVF PLAPSSGGTA ALGCLVKDYF P EPVTVSWN SGALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KVDKKVEPK |
-Macromolecule #6: VRC03 Light chain
| Macromolecule | Name: VRC03 Light chain / type: protein_or_peptide / ID: 6 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.043652 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPGI LSLSPGETAT LFCKASQGGN AMTWYQKRRG QVPRLLIYDT SRRASGVPDR FVGSGSGTDF FLTINKLDRE DFAVYYCQQ FEFFGLGSEL EVHRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL QSGNSQESVT E QDSKDSTY ...String: EIVLTQSPGI LSLSPGETAT LFCKASQGGN AMTWYQKRRG QVPRLLIYDT SRRASGVPDR FVGSGSGTDF FLTINKLDRE DFAVYYCQQ FEFFGLGSEL EVHRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL QSGNSQESVT E QDSKDSTY SLSSTLTLSK ADYEKHKVYA CEVTHQGLSS PVTKSFNRGE |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.01 / Component - Concentration: 150.0 mM / Component - Name: HEPES |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 298 K / Instrument: SPOTITON Details: 0.005% dodecyl maltoside was added to sample before vitification. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2206 / Average exposure time: 10.0 sec. / Average electron dose: 63.84 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6e5p: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)