[English] 日本語
 Yorodumi
Yorodumi- EMDB-0263: Horse spleen apo-ferritin with beam tilt correction in RELION-3.0 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0263 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Horse spleen apo-ferritin with beam tilt correction in RELION-3.0 | |||||||||
 Map data Map data | Reconstruction calculated from images that were collected over 9 Quantifoil holes using beam shift instead of moving the stage, and then correct for beam tilt in RELION-3.0 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
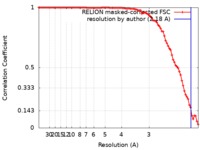
| Method | single particle reconstruction / cryo EM / Resolution: 2.18 Å | |||||||||
 Authors Authors | Zivanov J / Nakane T / Hagen WJH / Scheres SHW | |||||||||
 Citation Citation |  Journal: Elife / Year: 2018 Journal: Elife / Year: 2018Title: New tools for automated high-resolution cryo-EM structure determination in RELION-3. Authors: Jasenko Zivanov / Takanori Nakane / Björn O Forsberg / Dari Kimanius / Wim Jh Hagen / Erik Lindahl / Sjors Hw Scheres /    Abstract: Here, we describe the third major release of RELION. CPU-based vector acceleration has been added in addition to GPU support, which provides flexibility in use of resources and avoids memory ...Here, we describe the third major release of RELION. CPU-based vector acceleration has been added in addition to GPU support, which provides flexibility in use of resources and avoids memory limitations. Reference-free autopicking with Laplacian-of-Gaussian filtering and execution of jobs from python allows non-interactive processing during acquisition, including 2D-classification, model generation and 3D-classification. Per-particle refinement of CTF parameters and correction of estimated beam tilt provides higher resolution reconstructions when particles are at different heights in the ice, and/or coma-free alignment has not been optimal. Ewald sphere curvature correction improves resolution for large particles. We illustrate these developments with publicly available data sets: together with a Bayesian approach to beam-induced motion correction it leads to resolution improvements of 0.2-0.7 Å compared to previous RELION versions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0263.map.gz emd_0263.map.gz | 55.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0263-v30.xml emd-0263-v30.xml emd-0263.xml emd-0263.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0263_fsc.xml emd_0263_fsc.xml | 9 KB | Display |  FSC data file FSC data file |
| Images |  emd_0263.png emd_0263.png | 162.8 KB | ||
| Masks |  emd_0263_msk_1.map emd_0263_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_0263_half_map_1.map.gz emd_0263_half_map_1.map.gz emd_0263_half_map_2.map.gz emd_0263_half_map_2.map.gz | 47 MB 46.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0263 http://ftp.pdbj.org/pub/emdb/structures/EMD-0263 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0263 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0263 | HTTPS FTP |
-Validation report
| Summary document |  emd_0263_validation.pdf.gz emd_0263_validation.pdf.gz | 363.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0263_full_validation.pdf.gz emd_0263_full_validation.pdf.gz | 362.9 KB | Display | |
| Data in XML |  emd_0263_validation.xml.gz emd_0263_validation.xml.gz | 14.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0263 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0263 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0263 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0263 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0263.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0263.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction calculated from images that were collected over 9 Quantifoil holes using beam shift instead of moving the stage, and then correct for beam tilt in RELION-3.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
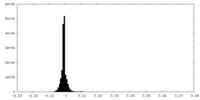
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_0263_msk_1.map emd_0263_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
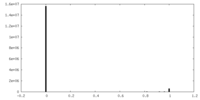
| Density Histograms |
-Half map: Independently refined halfmap 2
| File | emd_0263_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Independently refined halfmap 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
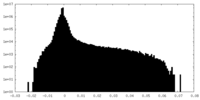
| Density Histograms |
-Half map: Independently refined halfmap 1
| File | emd_0263_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Independently refined halfmap 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
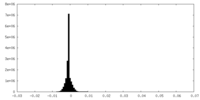
| Density Histograms |
- Sample components
Sample components
-Entire : Horse spleen apo-ferritin
| Entire | Name: Horse spleen apo-ferritin |
|---|---|
| Components |
|
-Supramolecule #1: Horse spleen apo-ferritin
| Supramolecule | Name: Horse spleen apo-ferritin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: horse spleen apo-ferritin
| Macromolecule | Name: horse spleen apo-ferritin / type: protein_or_peptide / ID: 1 / Details: Bought from Sigma / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MSSQIRQNYS TEVEAAVNRL VNLYLRASYT YLSLGFYFDR DDVALEGVCH FFRELAEEKR EGAERLLKMQ NQRGGRALFQ DLQKPSQDEW GTTLDAMKAA IVLEKSLNQA LLDLHALGSA QADPHLCDFL ESHFLDEEVK LIKKMGDHLT NIQRLVGSQA GLGEYLFERL TLKHD |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Material: COPPER |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Alignment procedure | Basic - Residual tilt: 1.2 mrad |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Details | Used beam shift to collect data, corrected for beam tilt in RELION-3.0 |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 450 / Average exposure time: 14.0 sec. / Average electron dose: 41.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 4.6 µm / Calibrated defocus min: 0.5 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)