1NHC
 
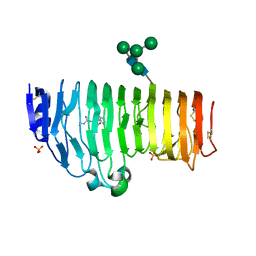 | | Structural insights into the processivity of endopolygalacturonase I from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | van Pouderoyen, G, Snijder, H.J, Benen, J.A, Dijkstra, B.W. | | Deposit date: | 2002-12-19 | | Release date: | 2003-11-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the processivity of endopolygalacturonase I from Aspergillus niger.
Febs Lett., 554, 2003
|
|
1I6W
 
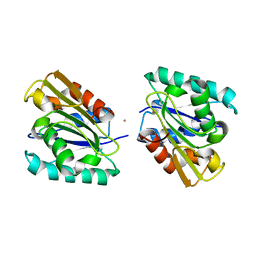 | | THE CRYSTAL STRUCTURE OF BACILLUS SUBTILIS LIPASE: A MINIMAL ALPHA/BETA HYDROLASE ENZYME | | Descriptor: | CADMIUM ION, LIPASE A | | Authors: | van Pouderoyen, G, Eggert, T, Jaeger, K.-E, Dijkstra, B.W. | | Deposit date: | 2001-03-05 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of Bacillus subtilis lipase: a minimal alpha/beta hydrolase fold enzyme.
J.Mol.Biol., 309, 2001
|
|
1TC3
 
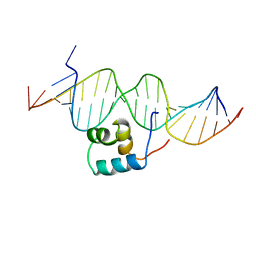 | | TRANSPOSASE TC3A1-65 FROM CAENORHABDITIS ELEGANS | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*GP*TP*CP*CP*TP*AP*TP*AP*GP*A P*AP*CP*TP*T)-3'), DNA (5'-D(*AP*GP*TP*TP*CP*TP*AP*TP*AP*GP*GP*AP*CP*CP*CP*CP*C P*CP*CP*T)-3'), PROTEIN (TC3 TRANSPOSASE) | | Authors: | Van Pouderoyen, G, Ketting, R.F, Perrakis, A, Plasterk, R.H.A, Sixma, T.K. | | Deposit date: | 1997-07-07 | | Release date: | 1997-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the specific DNA-binding domain of Tc3 transposase of C.elegans in complex with transposon DNA.
EMBO J., 16, 1997
|
|
3JUR
 
 | | The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima | | Descriptor: | Exo-poly-alpha-D-galacturonosidase | | Authors: | Pijning, T, van Pouderoyen, G, Kluskens, L.D, van der Oost, J, Dijkstra, B.W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The crystal structure of a hyperthermoactive exopolygalacturonase from Thermotoga maritima reveals a unique tetramer
Febs Lett., 2009
|
|
1U78
 
 | |
1YOB
 
 | | C69A Flavodoxin II from Azotobacter vinelandii | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin 2, SULFATE ION | | Authors: | Alagaratnam, S, van Pouderoyen, G, Pijning, T, Dijkstra, B.W, Cavazzini, D, Rossi, G.L, Canters, G.W. | | Deposit date: | 2005-01-27 | | Release date: | 2005-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A crystallographic study of Cys69Ala flavodoxin II from Azotobacter vinelandii: structural determinants of redox potential
Protein Sci., 14, 2005
|
|
1R50
 
 | | Bacillus subtilis lipase A with covalently bound Sc-IPG-phosphonate-inhibitor | | Descriptor: | Lipase, [(4S)-2,2-DIMETHYL-1,3-DIOXOLAN-4-YL]METHYL HYDROGEN HEX-5-ENYLPHOSPHONATE | | Authors: | Droege, M.J, Van Pouderoyen, G, Vrenken, T.E, Rueggeberg, C.J, Reetz, M.T, Dijkstra, B.W, Quax, W.J. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Directed Evolution of Bacillus subtilis Lipase A by Use of Enantiomeric Phosphonate Inhibitors: Crystal Structures and Phage Display Selection
Chembiochem, 7, 2005
|
|
1R4Z
 
 | | Bacillus subtilis lipase A with covalently bound Rc-IPG-phosphonate-inhibitor | | Descriptor: | Lipase, [(4R)-2,2-DIMETHYL-1,3-DIOXOLAN-4-YL]METHYL HYDROGEN HEX-5-ENYLPHOSPHONATE | | Authors: | Droege, M.J, Van Pouderoyen, G, Vrenken, T.E, Rueggeberg, C.J, Reetz, M.T, Dijkstra, B.W, Quax, W.J. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed Evolution of Bacillus subtilis Lipase A by Use of Enantiomeric Phosphonate Inhibitors: Crystal Structures and Phage Display Selection
Chembiochem, 7, 2005
|
|
2HIH
 
 | | Crystal structure of Staphylococcus hyicus lipase | | Descriptor: | CALCIUM ION, Lipase 46 kDa form, ZINC ION | | Authors: | Tiesinga, J.J.W, van Pouderoyen, G, Nardini, M, Dijkstra, B.W. | | Deposit date: | 2006-06-29 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis of phospholipase activity of Staphylococcus hyicus lipase.
J.Mol.Biol., 371, 2007
|
|
