5GY6
 
 | | Ribonuclease from Hericium erinaceus (RNase He1) | | Descriptor: | Ribonuclease T1, ZINC ION | | Authors: | Kobayashi, H, Sangawa, T, Takebe, K, Itagaki, T, Motoyoshi, N, Suzuki, M. | | Deposit date: | 2016-09-21 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ribonuclease from Hericium erinaceus (RNase He1)
To Be Published
|
|
3WQB
 
 | | Crystal structure of aeromonas sobria serine protease (ASP) and the chaperone (ORF2) complex | | Descriptor: | CALCIUM ION, Extracellular serine protease, Open reading frame 2 | | Authors: | Kobayashi, H, Yoshida, T, Miyakawa, T, Kato, R, Tashiro, M, Yamanaka, H, Tanokura, M, Tsuge, H. | | Deposit date: | 2014-01-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural Basis for Action of the External Chaperone for a Propeptide-deficient Serine Protease from Aeromonas sobria.
J.Biol.Chem., 290, 2015
|
|
3WHO
 
 | | X-ray-Crystallographic Structure of an RNase Po1 Exhibiting Anti-tumor Activity | | Descriptor: | Guanyl-specific ribonuclease Po1 | | Authors: | Kobayashi, H, Katsurtani, T, Hara, Y, Motoyoshi, N, Itagaki, T, Akita, F, Higashiura, A, Yamada, Y, Suzuki, M, Inokuchi, N. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystallographic structure of RNase Po1 that exhibits anti-tumor activity.
Biol.Pharm.Bull., 37, 2014
|
|
3HJR
 
 | | Crystal structure of serine protease of Aeromonas sobria | | Descriptor: | CALCIUM ION, Extracellular serine protease | | Authors: | Utsunomiya, H, Tsuge, H, Kobayashi, H, Okamoto, K. | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the kexin-like serine protease from Aeromonas sobria as a sepsis-causing factor
J.Biol.Chem., 284, 2009
|
|
2MK4
 
 | | Solution structure of ORF2 | | Descriptor: | Open reading frame 2 | | Authors: | Miyakawa, T, Kobayashi, H, Tashiro, M, Yamanaka, H, Tanokura, M. | | Deposit date: | 2014-01-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Action of the External Chaperone for a Propeptide-deficient Serine Protease from Aeromonas sobria.
J.Biol.Chem., 290, 2015
|
|
1IT0
 
 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, endo-1,4-beta-D-xylanase | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
1ISV
 
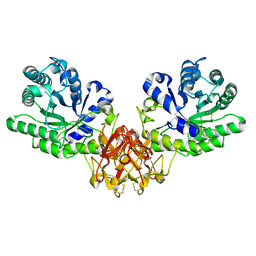 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylose | | Descriptor: | beta-D-xylopyranose, endo-1,4-beta-D-xylanase | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
1ISW
 
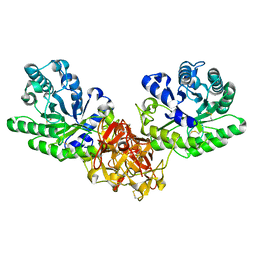 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylobiose | | Descriptor: | beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, endo-1,4-beta-D-xylanase | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
1ISZ
 
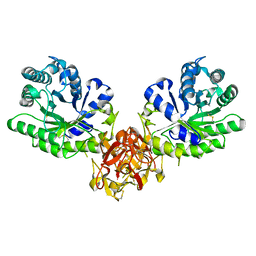 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with galactose | | Descriptor: | beta-D-galactopyranose, endo-1,4-beta-D-xylanase | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
1ISX
 
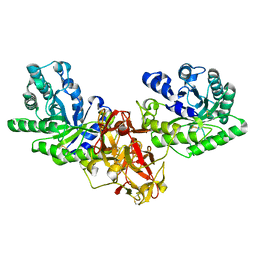 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylotriose | | Descriptor: | beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
1ISY
 
 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with glucose | | Descriptor: | beta-D-glucopyranose, endo-1,4-beta-D-xylanase | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
3A5V
 
 | | Crystal structure of alpha-galactosidase I from Mortierella vinacea | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fujimoto, Z, Kaneko, S, Kobayashi, H. | | Deposit date: | 2009-08-12 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Tetramer Structure of the Glycoside Hydrolase Family 27 alpha-Galactosidase I from Umbelopsis vinacea
Biosci.Biotechnol.Biochem., 73, 2009
|
|
7W05
 
 | | 12 mutant Ribonuclease from Hericium erinaceus GMP binding form | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE, Ribonuclease T1 | | Authors: | Takebe, K, Chida, T, Suzuki, M, Itagaki, T, Morita, Y, Uzawa, N, Kobayashi, H. | | Deposit date: | 2021-11-17 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | 12 mutant Ribonuclease from Hericium erinaceus GMP binding form
To Be Published
|
|
3WR2
 
 | |
1UAS
 
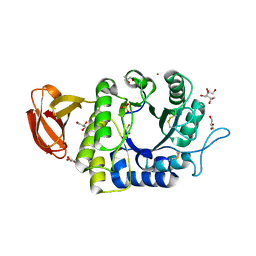 | | Crystal structure of rice alpha-galactosidase | | Descriptor: | GLYCEROL, PLATINUM (II) ION, SULFATE ION, ... | | Authors: | Fujimoto, Z, Kaneko, S, Momma, M, Kobayashi, H, Mizuno, H. | | Deposit date: | 2003-03-18 | | Release date: | 2003-07-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of rice alpha-galactosidase complexed with D-galactose
J.Biol.Chem., 278, 2003
|
|
6LS1
 
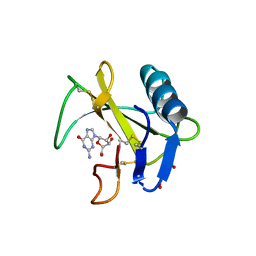 | | Ribonuclease from Hericium erinaceus active and GMP binding form | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE, Ribonuclease T1, ... | | Authors: | Takebe, K, Suzuki, M, Sangawa, T, Kobayashi, H, Itagaki, T. | | Deposit date: | 2020-01-16 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Ribonuclease from Hericium erinaceus active and GMP binding form
To Be Published
|
|
6LS8
 
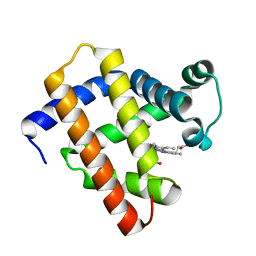 | | The monomeric structure of G80A/H81A/H82A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
6LTM
 
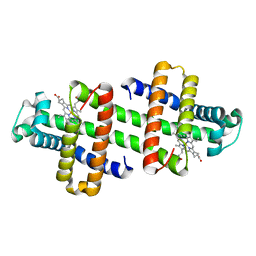 | | The dimeric structure of G80A/H81A/H82A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
6LTL
 
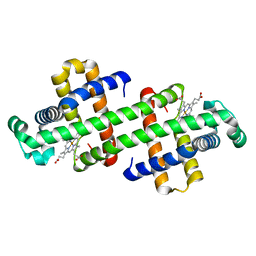 | | The dimeric structure of G80A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
1V6Y
 
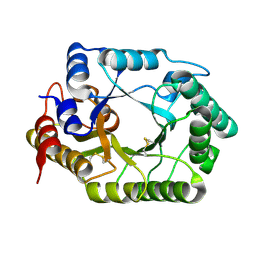 | | Crystal Structure Of chimeric Xylanase between Streptomyces Olivaceoviridis E-86 FXYN and Cellulomonas fimi Cex | | Descriptor: | Beta-xylanase,Exoglucanase/xylanase | | Authors: | Kaneko, S, Ichinose, H, Fujimoto, Z, Kuno, A, Yura, K, Go, M, Mizuno, H, Kusakabe, I, Kobayashi, H. | | Deposit date: | 2003-12-04 | | Release date: | 2004-09-07 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of a family 10 beta-xylanase chimera of Streptomyces olivaceoviridis E-86 FXYN and Cellulomonas fimi Cex
J.Biol.Chem., 279, 2004
|
|
1WR1
 
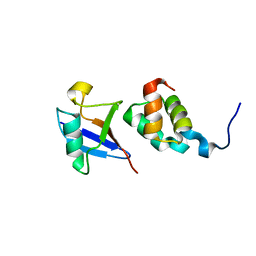 | | The complex structure of Dsk2p UBA with ubiquitin | | Descriptor: | Ubiquitin, Ubiquitin-like protein DSK2 | | Authors: | Ohno, A, Jee, J.G, Fujiwara, K, Tenno, T, Goda, N, Tochio, H, Hiroaki, H, kobayashi, H, Shirakawa, M. | | Deposit date: | 2004-10-08 | | Release date: | 2005-04-19 | | Last modified: | 2023-09-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the UBA domain of Dsk2p in complex with ubiquitin molecular determinants for ubiquitin recognition.
Structure, 13, 2005
|
|
1UAW
 
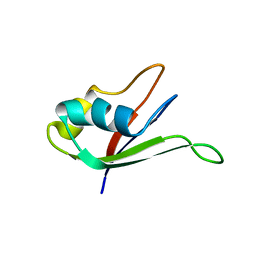 | | Solution structure of the N-terminal RNA-binding domain of mouse Musashi1 | | Descriptor: | mouse-musashi-1 | | Authors: | Miyanoiri, Y, Kobayashi, H, Watanabe, M, Ikeda, T, Nagata, T, Okano, H, Uesugi, S, Katahira, M. | | Deposit date: | 2003-03-24 | | Release date: | 2004-03-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Origin of higher affinity to RNA of the N-terminal RNA-binding domain than that of the C-terminal one of a mouse neural protein, musashi1, as revealed by comparison of their structures, modes of interaction, surface electrostatic potentials, and backbone dynamics
J.Biol.Chem., 278, 2003
|
|
1V6X
 
 | | Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 3(3)-4-O-methyl-alpha-D-glucuronosyl-xylotriose | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ENDO-1,4-BETA-D-XYLANASE, beta-D-xylopyranose, ... | | Authors: | Fujimoto, Z, Kaneko, S, Kuno, A, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2003-12-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86
J.Biol.Chem., 279, 2004
|
|
1V6V
 
 | | Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 3(2)-alpha-L-arabinofuranosyl-xylotriose | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, alpha-L-arabinofuranose-(1-3)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, beta-D-xylopyranose, ... | | Authors: | Fujimoto, Z, Kaneko, S, Kuno, A, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2003-12-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86
J.Biol.Chem., 279, 2004
|
|
1V6U
 
 | | Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-alpha-L-arabinofuranosyl-xylobiose | | Descriptor: | alpha-D-xylopyranose, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, beta-D-xylopyranose, ... | | Authors: | Fujimoto, Z, Kaneko, S, Kuno, A, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2003-12-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86
J.Biol.Chem., 279, 2004
|
|
