5Y51
 
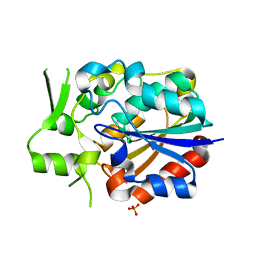 | | Crystal structure of PytH_H230A | | Descriptor: | Pyrethroid hydrolase, SULFATE ION | | Authors: | Xu, D.Q, Ran, T.T, He, J, Wang, W.W. | | Deposit date: | 2017-08-06 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Catalytic Mechanism of a Novel Pyrethroid Hydrolase from Sphingobium faniae JZ-2
To Be Published
|
|
5Y57
 
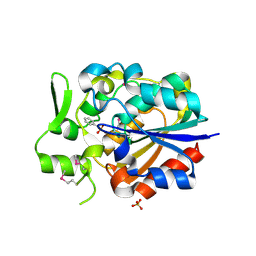 | |
5Y5V
 
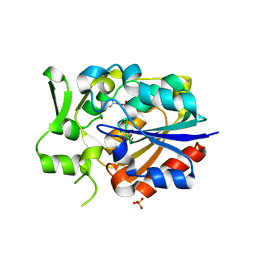 | |
5Y5R
 
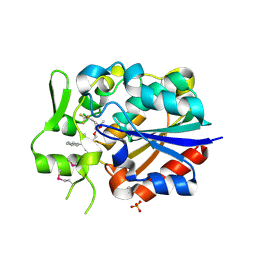 | | Crystal structure of a novel Pyrethroid Hydrolase PytH with BIF | | Descriptor: | (2-methyl-3-phenyl-phenyl)methyl (1~{S})-3-[(~{E})-2-chloranyl-3,3,3-tris(fluoranyl)prop-1-enyl]-2,2-dimethyl-cyclopropane-1-carboxylate, Pyrethroid hydrolase, SULFATE ION | | Authors: | Xu, D.Q, Ran, T.T, Wang, W.W. | | Deposit date: | 2017-08-09 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure and Catalytic Mechanism of a Novel Pyrethroid Hydrolase from Sphingobium faniae JZ-2
To Be Published
|
|
5ZJK
 
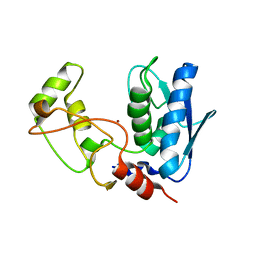 | | Structure of myroilysin | | Descriptor: | Myroilysin, PHOSPHATE ION, ZINC ION | | Authors: | Li, W.D, Ran, T.T, Xu, D.Q, Wang, W.W. | | Deposit date: | 2018-03-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of mature myroilysin and implication for its activation mechanism.
Int.J.Biol.Macromol., 2019
|
|
5HXK
 
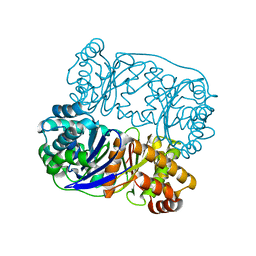 | |
5X1R
 
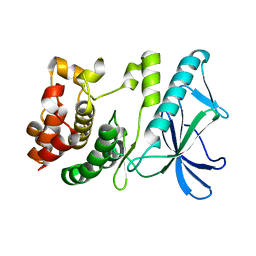 | | PpkA-294 apo form | | Descriptor: | PpkA-294 | | Authors: | Li, P.P, Ran, T.T, Xu, D.Q, Wang, W.W. | | Deposit date: | 2017-01-26 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the kinase domain of PpkA, a key regulatory component of T6SS, reveal a general inhibitory mechanism.
Biochem.J., 475, 2018
|
|
5X1T
 
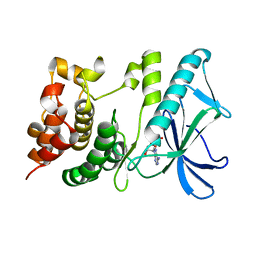 | | PpkA-294 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PpkA-294 | | Authors: | Li, P.P, Ran, T.T, Xu, D.Q, Wang, W.W. | | Deposit date: | 2017-01-26 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of the kinase domain of PpkA, a key regulatory component of T6SS, reveal a general inhibitory mechanism.
Biochem.J., 475, 2018
|
|
5X1Q
 
 | | PpkA-294 with ATP and MnCl2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Li, P.P, Ran, T.T, Xu, D.Q, Wang, W.W. | | Deposit date: | 2017-01-26 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Crystal structures of the kinase domain of PpkA, a key regulatory component of T6SS, reveal a general inhibitory mechanism.
Biochem.J., 475, 2018
|
|
5X1S
 
 | | PpkA-294 with Amppcp | | Descriptor: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, PpkA | | Authors: | Li, P.P, Ran, T.T, Xu, D.Q, Wang, W.W. | | Deposit date: | 2017-01-26 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of the kinase domain of PpkA, a key regulatory component of T6SS, reveal a general inhibitory mechanism.
Biochem.J., 475, 2018
|
|
4PPM
 
 | | Crystal structure of PigE: a transaminase involved in the biosynthesis of 2-methyl-3-n-amyl-pyrrole (MAP) from Serratia sp. FS14 | | Descriptor: | Aminotransferase, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Lou, X.D, Ran, T.T, Xu, D.Q, Wang, W.W. | | Deposit date: | 2014-02-27 | | Release date: | 2015-01-14 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of PigE: a transaminase involved in the biosynthesis of 2-methyl-3-n-amyl-pyrrole (MAP) from Serratia sp. FS14
Biochem.Biophys.Res.Commun., 447, 2014
|
|
5HNV
 
 | | Crystal structure of PpkA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Li, P.P, Ran, T.T, Wang, W.W. | | Deposit date: | 2016-01-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structures of the kinase domain of PpkA, a key regulatory component of T6SS, reveal a general inhibitory mechanism.
Biochem.J., 475, 2018
|
|
5D7W
 
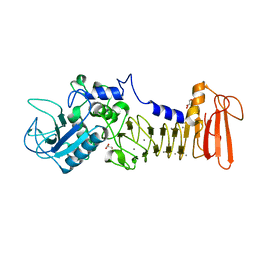 | | Crystal structure of serralysin | | Descriptor: | CALCIUM ION, GLYCEROL, Serralysin, ... | | Authors: | Wu, D, Ran, T, Xu, D.Q, Wang, W. | | Deposit date: | 2015-08-14 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of a thermostable serralysin from Serratia sp. FS14 at 1.1 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4DBH
 
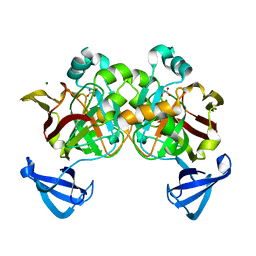 | | Crystal structure of Cg1458 with inhibitor | | Descriptor: | 2-HYDROXYHEPTA-2,4-DIENE-1,7-DIOATE ISOMERASE, MAGNESIUM ION, OXALATE ION | | Authors: | Ran, T.T, Wang, W.W, Xu, D.Q, Gao, Y.Y. | | Deposit date: | 2012-01-15 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structures of Cg1458 reveal a catalytic lid domain and a common catalytic mechanism for FAH family.
Biochem.J., 449, 2013
|
|
4DBF
 
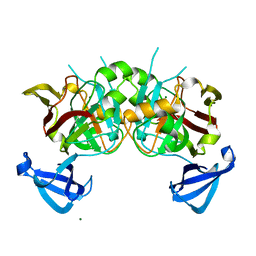 | | Crystal structures of Cg1458 | | Descriptor: | 2-HYDROXYHEPTA-2,4-DIENE-1,7-DIOATE ISOMERASE, MAGNESIUM ION | | Authors: | Ran, T.T, Xu, D.Q, Wang, W.W, Gao, Y.Y, Wang, M.T. | | Deposit date: | 2012-01-15 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Cg1458 reveal a catalytic lid domain and a common catalytic mechanism for FAH family.
Biochem.J., 449, 2013
|
|
6AEO
 
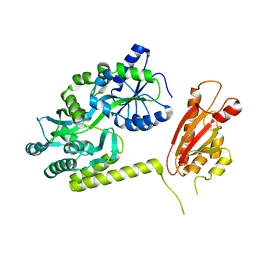 | | TssL periplasmic domain | | Descriptor: | GLYCEROL, Maltose/maltodextrin-binding periplasmic protein,TssL | | Authors: | Ran, T.T, Wang, W.W, Wang, X.B, Xu, D.Q. | | Deposit date: | 2018-08-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the periplasmic domain of TssL, a key membrane component of Type VI secretion system.
Int.J.Biol.Macromol., 120, 2018
|
|
