1AUL
 
 | | SOLUTION STRUCTURE OF A HIGHLY STABLE DNA DUPLEX CONJUGATED TO A MINOR GROOVE BINDER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*TP*AP*AP*TP*CP*A)-3'), DNA (5'-D(P*THXP*GP*AP*TP*TP*AP*TP*CP*TP*G)-3') | | Authors: | Kumar, S, Reed, M.W, Gamper Junior, H.B, Gorn, V.V, Lukhtanov, E.A, Foti, M, West, J, Meyer Junior, R.B, Schweitzer, B.I. | | Deposit date: | 1997-08-29 | | Release date: | 1997-12-24 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a highly stable DNA duplex conjugated to a minor groove binder.
Nucleic Acids Res., 26, 1998
|
|
1JBL
 
 | | Solution structure of SFTI-1, A cyclic trypsin inhibitor from sunflower seeds | | Descriptor: | CYCLIC TRYPSIN INHIBITOR | | Authors: | Korsinczky, M.L.J, Schirra, H.J, Rosengren, K.J, West, J, Condie, B.A, Otvos, L, Anderson, M.A, Craik, D.J. | | Deposit date: | 2001-06-05 | | Release date: | 2001-08-22 | | Last modified: | 2015-04-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures by 1H NMR of the novel cyclic trypsin inhibitor SFTI-1 from sunflower seeds and an acyclic permutant.
J.Mol.Biol., 311, 2001
|
|
1JBN
 
 | | Solution structure of an acyclic permutant of SFTI-1, A trypsin inhibitor from sunflower seeds | | Descriptor: | CYCLIC TRYPSIN INHIBITOR | | Authors: | Korsinczky, M.L.J, Schirra, H.J, Rosengren, K.J, West, J, Condie, B.A, Otvos, L, Anderson, M.A, Craik, D.J. | | Deposit date: | 2001-06-06 | | Release date: | 2001-08-22 | | Last modified: | 2016-12-28 | | Method: | SOLUTION NMR | | Cite: | Solution structures by 1H NMR of the novel cyclic trypsin inhibitor SFTI-1 from sunflower seeds and an acyclic permutant.
J.Mol.Biol., 311, 2001
|
|
1MI2
 
 | | SOLUTION STRUCTURE OF MURINE MACROPHAGE INFLAMMATORY PROTEIN-2, NMR, 20 STRUCTURES | | Descriptor: | MACROPHAGE INFLAMMATORY PROTEIN-2 | | Authors: | Shao, W, Jerva, L.F, West, J, Lolis, E, Schweitzer, B.I. | | Deposit date: | 1997-10-24 | | Release date: | 1998-04-29 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of murine macrophage inflammatory protein-2.
Biochemistry, 37, 1998
|
|
8S2W
 
 | |
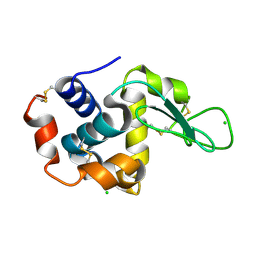
8S2U
 
 | |
8S2V
 
 | |
8S2X
 
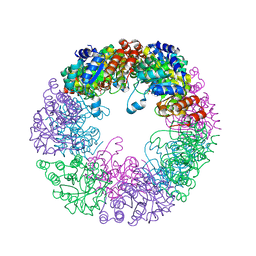 | |
5OW6
 
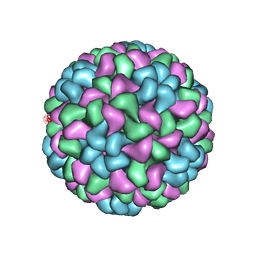 | | CryoEM structure of recombinant CMV particles with Tetanus-epitope | | Descriptor: | Capsid protein, VP2, VP3, ... | | Authors: | Kotecha, A, Stuart, D.I, Backmann, M. | | Deposit date: | 2017-08-30 | | Release date: | 2017-09-13 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Incorporation of tetanus-epitope into virus-like particles achieves vaccine responses even in older recipients in models of psoriasis, Alzheimer's and cat allergy.
NPJ Vaccines, 2, 2017
|
|
6EUS
 
 | |
5SZD
 
 | | Crystal structure of Aquifex aeolicus Hfq at 1.5A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Stanek, K, Patterson, J, Randolph, P.S, Mura, C. | | Deposit date: | 2016-08-13 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.494 Å) | | Cite: | Crystal structure and RNA-binding properties of an Hfq homolog from the deep-branching Aquificae: conservation of the lateral RNA-binding mode.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5SZE
 
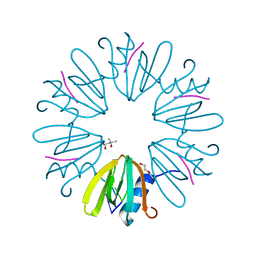 | | Crystal structure of Aquifex aeolicus Hfq-RNA complex at 1.5A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, DI(HYDROXYETHYL)ETHER, RNA (5'-R(P*UP*UP*U)-3'), ... | | Authors: | Stanek, K, Patterson, J, Randolph, P.S, Mura, C. | | Deposit date: | 2016-08-13 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure and RNA-binding properties of an Hfq homolog from the deep-branching Aquificae: conservation of the lateral RNA-binding mode.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1HHV
 
 | | SOLUTION STRUCTURE OF VIRUS CHEMOKINE VMIP-II | | Descriptor: | VIRUS CHEMOKINE VMIP-II | | Authors: | Shao, W, Fernandez, E, Navenot, J.M, Wilken, J, Thompson, D.A, Pepiper, S, Schweitzer, B.I, Lolis, E. | | Deposit date: | 1998-12-06 | | Release date: | 2003-09-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | CCR2 and CCR5 receptor-binding properties of herpesvirus-8
vMIP-II based on sequence analysis and its solution structure
Eur.J.Biochem., 268, 2001
|
|
