8JP6
 
 | |
8JPG
 
 | | Cryo-EM structure of full-length ERGIC-53 with MCFD2 | | Descriptor: | CALCIUM ION, Multiple coagulation factor deficiency protein 2, Protein ERGIC-53, ... | | Authors: | Watanabe, S, Inaba, K. | | Deposit date: | 2023-06-12 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (6.76 Å) | | Cite: | Structure of full-length ERGIC-53 in complex with MCFD2 for cargo transport.
Nat Commun, 15, 2024
|
|
8JP8
 
 | |
8JP4
 
 | |
8JP7
 
 | |
8JP9
 
 | |
8JP5
 
 | |
5GU7
 
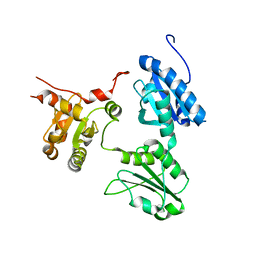 | | Crystal Structure of human ERp44 form II | | Descriptor: | Endoplasmic reticulum resident protein 44 | | Authors: | Watanabe, S, Inaba, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of pH-dependent client binding by ERp44, a key regulator of protein secretion at the ER-Golgi interface
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5GU6
 
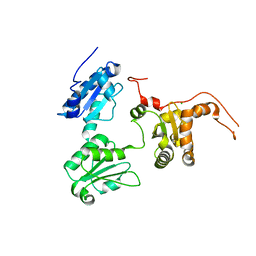 | | Crystal structure of Human ERp44 form I | | Descriptor: | CHLORIDE ION, Endoplasmic reticulum resident protein 44 | | Authors: | Watanabe, S, Inaba, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of pH-dependent client binding by ERp44, a key regulator of protein secretion at the ER-Golgi interface
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XWM
 
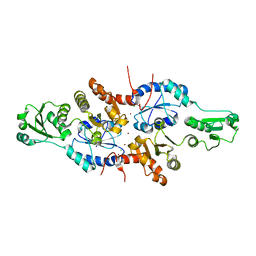 | | human ERp44 zinc-bound form | | Descriptor: | CHLORIDE ION, Endoplasmic reticulum resident protein 44, ZINC ION | | Authors: | Watanabe, S, Harayama, M, Inaba, K. | | Deposit date: | 2017-06-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Zinc regulates ERp44-dependent protein quality control in the early secretory pathway.
Nat Commun, 10, 2019
|
|
5AUP
 
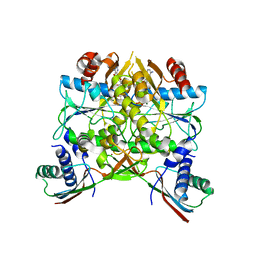 | | Crystal structure of the HypAB complex | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUN
 
 | | Crystal structure of the HypAB-Ni complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase involved in chromosome partitioning, ParA/MinD family, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUO
 
 | | Crystal structure of the HypAB-Ni complex (AMPPCP) | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUQ
 
 | | Crystal structure of ATPase-type HypB in the nucleotide free state | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3VYT
 
 | | Crystal structure of the HypC-HypD-HypE complex (form I inward) | | Descriptor: | CHLORIDE ION, Hydrogenase expression/formation protein HypC, Hydrogenase expression/formation protein HypD, ... | | Authors: | Watanabe, S, Miki, K. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Structure, 20, 2012
|
|
3VYS
 
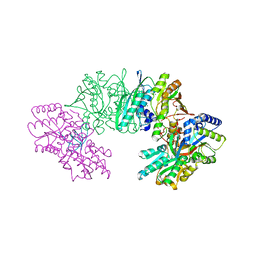 | | Crystal structure of the HypC-HypD-HypE complex (form I) | | Descriptor: | Hydrogenase expression/formation protein HypC, Hydrogenase expression/formation protein HypD, Hydrogenase expression/formation protein HypE, ... | | Authors: | Watanabe, S, Miki, K. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Structure, 20, 2012
|
|
3VYR
 
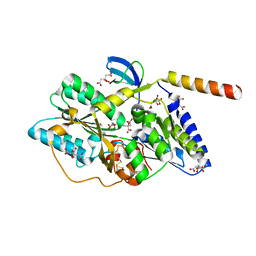 | | Crystal structure of the HypC-HypD complex | | Descriptor: | CITRIC ACID, Hydrogenase expression/formation protein HypC, Hydrogenase expression/formation protein HypD, ... | | Authors: | Watanabe, S, Miki, K. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Structure, 20, 2012
|
|
3VYU
 
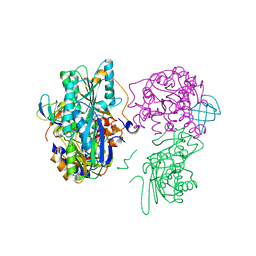 | | Crystal structure of the HypC-HypD-HypE complex (form II) | | Descriptor: | Hydrogenase expression/formation protein HypC, Hydrogenase expression/formation protein HypD, Hydrogenase expression/formation protein HypE, ... | | Authors: | Watanabe, S, Miki, K. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Structure, 20, 2012
|
|
2D2E
 
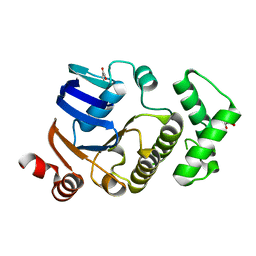 | | Crystal structure of atypical cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, GLYCEROL, SufC protein | | Authors: | Watanabe, S, Kita, A, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-09-08 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Atypical Cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8.
J.Mol.Biol., 353, 2005
|
|
2D2F
 
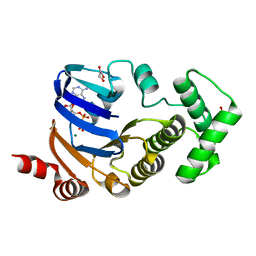 | | Crystal structure of atypical cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Watanabe, S, Kita, A, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-09-08 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Atypical Cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8.
J.Mol.Biol., 353, 2005
|
|
2Z1D
 
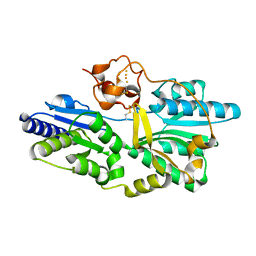 | | Crystal structure of [NiFe] hydrogenase maturation protein, HypD from Thermococcus kodakaraensis | | Descriptor: | Hydrogenase expression/formation protein hypD, IRON/SULFUR CLUSTER | | Authors: | Watanabe, S, Matsumi, R, Arai, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2007-05-08 | | Release date: | 2007-07-17 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structures of [NiFe] Hydrogenase Maturation Proteins HypC, HypD, and HypE: Insights into Cyanation Reaction by Thiol Redox Signaling
Mol.Cell, 27, 2007
|
|
2ZHH
 
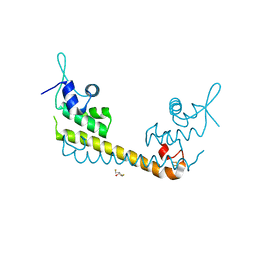 | | Crystal structure of SoxR | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, FE2/S2 (INORGANIC) CLUSTER, Redox-sensitive transcriptional activator soxR | | Authors: | Watanabe, S, Kita, A, Kobayashi, K, Miki, K. | | Deposit date: | 2008-02-05 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3A43
 
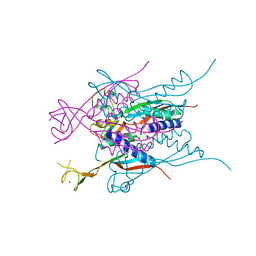 | | Crystal structure of HypA | | Descriptor: | Hydrogenase nickel incorporation protein hypA, ZINC ION | | Authors: | Watanabe, S, Arai, T, Matsumi, R, Aromi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-06-30 | | Release date: | 2009-10-06 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of HypA, a nickel-binding metallochaperone for [NiFe] hydrogenase maturation.
J.Mol.Biol., 394, 2009
|
|
3A44
 
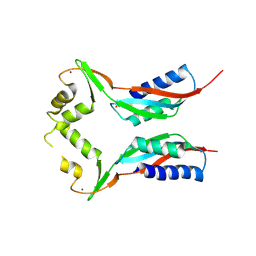 | | Crystal structure of HypA in the dimeric form | | Descriptor: | Hydrogenase nickel incorporation protein hypA, ZINC ION | | Authors: | Watanabe, S, Arai, T, Matsumi, R, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-06-30 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure of HypA, a nickel-binding metallochaperone for [NiFe] hydrogenase maturation.
J.Mol.Biol., 394, 2009
|
|
2ZHG
 
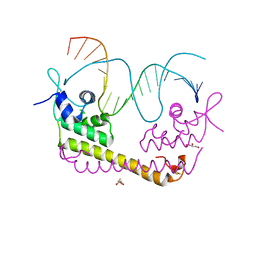 | | Crystal structure of SoxR in complex with DNA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA (5'-D(*DGP*DCP*DCP*DTP*DCP*DAP*DAP*DGP*DTP*DTP*DAP*DAP*DCP*DTP*DTP*DGP*DAP*DGP*DGP*DC)-3'), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Watanabe, S, Kita, A, Kobayashi, K, Miki, K. | | Deposit date: | 2008-02-05 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
