4DXW
 
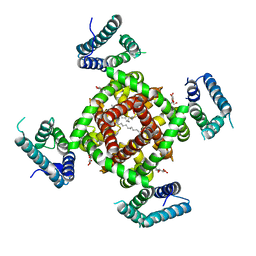 | | Crystal structure of NavRh, a voltage-gated sodium channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein, ... | | Authors: | Zhang, X, Ren, W.L, Yan, C.Y, Wang, J.W, Yan, N. | | Deposit date: | 2012-02-28 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel
Nature, 486, 2012
|
|
4F1Z
 
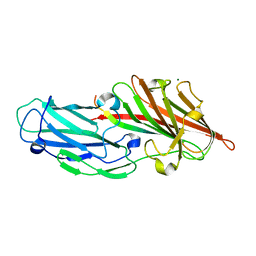 | | Crystal structures reveal the multi-ligand binding mechanism of the Staphylococcus aureus ClfB | | Descriptor: | Clumping factor B, MAGNESIUM ION, peptide from Keratin, ... | | Authors: | Yang, M.J, Xiang, H, Wang, J.W, Liu, B, Chen, Y.G, Liu, L, Deng, X.M, Feng, Y. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures Reveal the Multi-Ligand Binding Mechanism of Staphylococcus aureus ClfB
Plos Pathog., 8, 2012
|
|
4F24
 
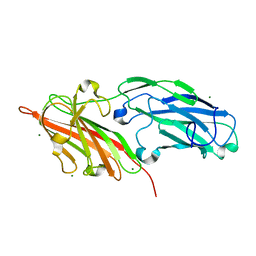 | | Crystal structures reveal the multi-ligand binding mechanism of the Staphylococcus aureus ClfB | | Descriptor: | Clumping factor B, MAGNESIUM ION | | Authors: | Yang, M.J, Xiang, H, Wang, J.W, Liu, B, Chen, Y.G, Liu, L, Deng, X.M, Feng, Y. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Crystal Structures Reveal the Multi-Ligand Binding Mechanism of Staphylococcus aureus ClfB
Plos Pathog., 8, 2012
|
|
4F27
 
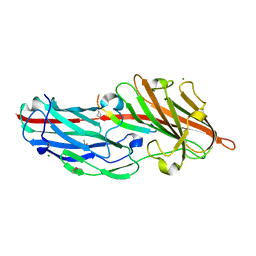 | | Crystal structures reveal the multi-ligand binding mechanism of the Staphylococcus aureus ClfB | | Descriptor: | Clumping factor B, MAGNESIUM ION, peptide from Fibrinogen alpha chain | | Authors: | Yang, M.J, Xiang, H, Wang, J.W, Liu, B, Chen, Y.G, Liu, L, Deng, X.M, Feng, Y. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.917 Å) | | Cite: | Crystal Structures Reveal the Multi-Ligand Binding Mechanism of Staphylococcus aureus ClfB
Plos Pathog., 8, 2012
|
|
4DJI
 
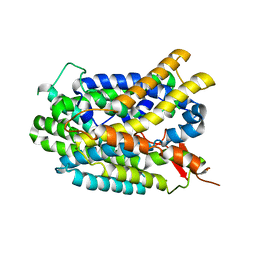 | | Structure of glutamate-GABA antiporter GadC | | Descriptor: | Probable glutamate/gamma-aminobutyrate antiporter | | Authors: | Ma, D, Lu, P.L, Yan, C.Y, Fan, C, Yin, P, Wang, J.W, Shi, Y.G. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.187 Å) | | Cite: | Structure and mechanism of a glutamate-GABA antiporter
Nature, 483, 2012
|
|
4DJK
 
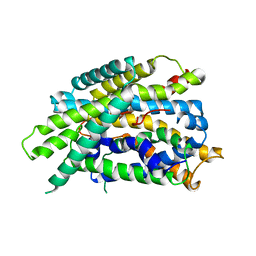 | | Structure of glutamate-GABA antiporter GadC | | Descriptor: | Probable glutamate/gamma-aminobutyrate antiporter | | Authors: | Ma, D, Lu, P.L, Yan, C.Y, Fan, C, Yin, P, Wang, J.W, Shi, Y.G. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structure and mechanism of a glutamate-GABA antiporter
Nature, 483, 2012
|
|
8XAT
 
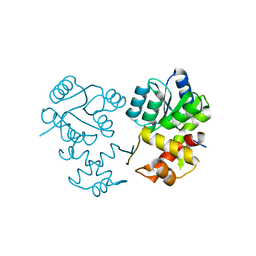 | |
8XAS
 
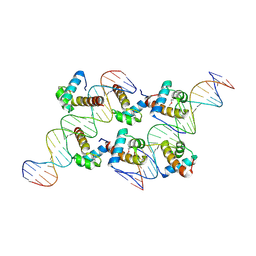 | | Crystal structure of AtARR1-DBD in complex with a DNA fragment | | Descriptor: | DNA (50-MER), Two-component response regulator ARR1 | | Authors: | Li, J.X, Zhou, C.M, zhang, P, Wang, J.W. | | Deposit date: | 2023-12-05 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | The structure of B-ARR reveals the molecular basis of transcriptional activation by cytokinin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4F20
 
 | | Crystal structures reveal the multi-ligand binding mechanism of the Staphylococcus aureus ClfB | | Descriptor: | Clumping factor B, MAGNESIUM ION, peptide from Dermokine | | Authors: | Yang, M.J, Xiang, H, Wang, J.W, Liu, B, Chen, Y.G, Liu, L, Deng, X.M, Feng, Y. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal Structures Reveal the Multi-Ligand Binding Mechanism of Staphylococcus aureus ClfB
Plos Pathog., 8, 2012
|
|
3V6T
 
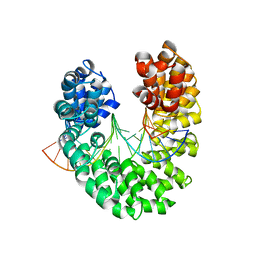 | | Crystal structure of the DNA-bound dHax3, a TAL effector, at 1.85 angstrom | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), dHax3 | | Authors: | Deng, D, Yan, C.Y, Pan, X.J, Wang, J.W, Yan, N, Shi, Y.G. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Sequence-Specific Recognition of DNA by TAL Effectors
Science, 2012
|
|
3V6P
 
 | | Crystal structure of the DNA-binding domain of dHax3, a TAL effector | | Descriptor: | dHax3 | | Authors: | Deng, D, Yan, C.Y, Pan, X.J, Wang, J.W, Shi, Y.G, Yan, N. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural Basis for Sequence-Specific Recognition of DNA by TAL Effectors
Science, 2012
|
|
3PZ7
 
 | |
3PXI
 
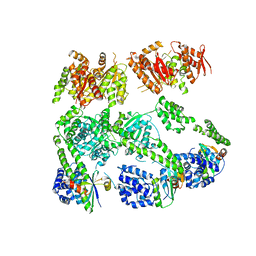 | | Structure of MecA108:ClpC | | Descriptor: | Adapter protein mecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | Deposit date: | 2010-12-09 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (6.926 Å) | | Cite: | Structure and mechanism of the hexameric MecA-ClpC molecular machine.
Nature, 471, 2011
|
|
6K1H
 
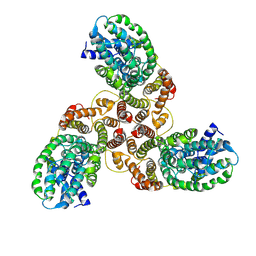 | | Structure of membrane protein | | Descriptor: | PTS mannose transporter subunit IID, PTS system mannose-specific EIIC component, alpha-D-mannopyranose | | Authors: | Wang, J.W, Zeng, J.W. | | Deposit date: | 2019-05-10 | | Release date: | 2019-07-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structure of the mannose transporter of the bacterial phosphotransferase system.
Cell Res., 29, 2019
|
|
7DYR
 
 | |
9J1J
 
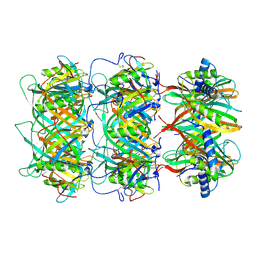 | | Cap region of monocin | | Descriptor: | AA protein, DUF5072 domain-containing protein | | Authors: | Wang, J.W, Gu, Z.W. | | Deposit date: | 2024-08-05 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structure of an F-type phage tail-like bacteriocin from Listeria monocytogenes.
Nat Commun, 16, 2025
|
|
9J1K
 
 | | Tip region of monocin | | Descriptor: | AA protein, CCA-adding enzyme, FtbJ, ... | | Authors: | Wang, J.W, Gu, Z.W. | | Deposit date: | 2024-08-05 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structure of an F-type phage tail-like bacteriocin from Listeria monocytogenes.
Nat Commun, 16, 2025
|
|
9J1L
 
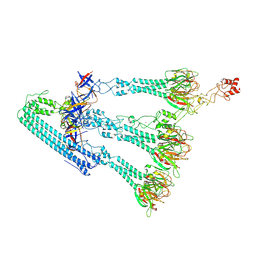 | | Side fiber of monocin | | Descriptor: | Alpha-amylase, FE (III) ION, FtbO, ... | | Authors: | Wang, J.W, Gu, Z.W. | | Deposit date: | 2024-08-05 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of an F-type phage tail-like bacteriocin from Listeria monocytogenes.
Nat Commun, 16, 2025
|
|
7VLY
 
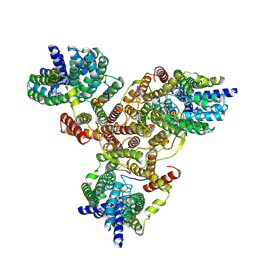 | | Cryo-EM structure of Listeria monocytogenes man-PTS complexed with pediocin PA-1 | | Descriptor: | Bacteriocin pediocin PA-1, Mannose/fructose/sorbose family PTS transporter subunit IIC, PTS mannose family transporter subunit IID, ... | | Authors: | Wang, J.W. | | Deposit date: | 2021-10-05 | | Release date: | 2021-12-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural Basis of Pore Formation in the Mannose Phosphotransferase System by Pediocin PA-1.
Appl.Environ.Microbiol., 88, 2022
|
|
7VLX
 
 | | Cryo-EM structures of Listeria monocytogenes man-PTS | | Descriptor: | Mannose/fructose/sorbose family PTS transporter subunit IIC, PTS mannose family transporter subunit IID, alpha-D-mannopyranose | | Authors: | Wang, J.W. | | Deposit date: | 2021-10-05 | | Release date: | 2021-12-01 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural Basis of Pore Formation in the Mannose Phosphotransferase System by Pediocin PA-1.
Appl.Environ.Microbiol., 88, 2022
|
|
7VI9
 
 | |
7VII
 
 | |
7VIA
 
 | |
7VIK
 
 | |
4RP9
 
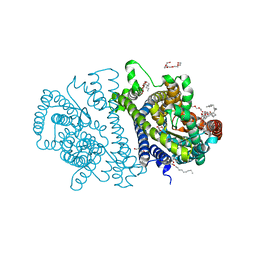 | | Bacterial vitamin C transporter UlaA/SgaT in C2 form | | Descriptor: | ASCORBIC ACID, Ascorbate-specific permease IIC component UlaA, PENTAETHYLENE GLYCOL, ... | | Authors: | Wang, J.W. | | Deposit date: | 2014-10-29 | | Release date: | 2015-03-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Crystal structure of a phosphorylation-coupled vitamin C transporter.
Nat.Struct.Mol.Biol., 22, 2015
|
|
