2G2L
 
 | | Crystal Structure of the Second PDZ Domain of SAP97 in Complex with a GluR-A C-terminal Peptide | | Descriptor: | 18-mer peptide from glutamate receptor, ionotropic, AMPA1, ... | | Authors: | Von Ossowski, I, Oksanen, E, Von Ossowski, L, Cai, C, Sundberg, M, Goldman, A, Keinanen, K. | | Deposit date: | 2006-02-16 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the second PDZ domain of SAP97 in complex with a GluR-A C-terminal peptide
Febs J., 273, 2006
|
|
2AWX
 
 | | Synapse associated protein 97 PDZ2 domain variant C378S | | Descriptor: | HISTIDINE, Synapse associated protein 97 | | Authors: | Von Ossowski, I, Oksanen, E, Von Ossowski, L, Cai, C, Sundberg, M, Goldman, A, Keinanen, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the second PDZ domain of SAP97 in complex with a GluR-A C-terminal peptide
Febs J., 273, 2006
|
|
2AWW
 
 | | Synapse associated protein 97 PDZ2 domain variant C378G with C-terminal GluR-A peptide | | Descriptor: | 18-residue C-terminal peptide from glutamate receptor, ionotropic, AMPA1, ... | | Authors: | Von Ossowski, I, Oksanen, E, Von Ossowski, L, Cai, C, Sundberg, M, Goldman, A, Keinanen, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the second PDZ domain of SAP97 in complex with a GluR-A C-terminal peptide
Febs J., 273, 2006
|
|
2AWU
 
 | | Synapse associated protein 97 PDZ2 domain variant C378G | | Descriptor: | AHH, Synapse-associated protein 97 | | Authors: | Von Ossowski, I, Oksanen, E, Von Ossowski, L, Cai, C, Sundberg, M, Goldman, A, Keinanen, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of the second PDZ domain of SAP97 in complex with a GluR-A C-terminal peptide
Febs J., 273, 2006
|
|
6M3Y
 
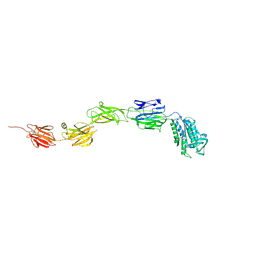 | | Crystal structure of pilus adhesin, SpaC from Lactobacillus rhamnosus GG - open conformation | | Descriptor: | MAGNESIUM ION, Pilus assembly protein | | Authors: | Kant, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2020-03-04 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of lactobacillar SpaC reveals an atypical five-domain pilus tip adhesin: Exposing its substrate-binding and assembly in SpaCBA pili.
J.Struct.Biol., 211, 2020
|
|
6M7C
 
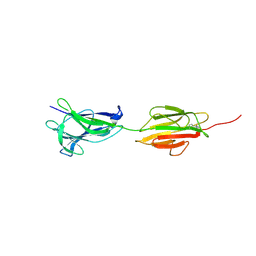 | |
8W5B
 
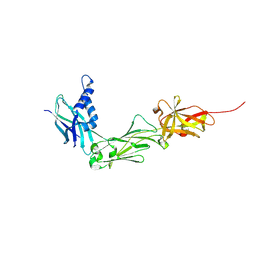 | | Crystal Structure of the shaft pilin LrpA from Ligilactobacillus ruminis | | Descriptor: | IODIDE ION, LPXTG-motif cell wall anchor domain protein | | Authors: | Prajapati, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2023-08-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The crystal structure of the N-terminal domain of the backbone pilin LrpA reveals a new closure-and-twist motion for assembling dynamic pili in Ligilactobacillus ruminis.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8WB8
 
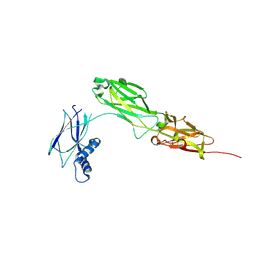 | |
7BVX
 
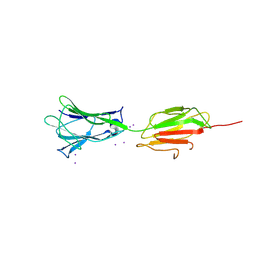 | | Crystal structure of C-terminal fragment of pilus adhesin SpaC from Lactobacillus rhamnosus GG-Iodide soaked | | Descriptor: | IODIDE ION, Pilus assembly protein | | Authors: | Kant, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2020-04-12 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Crystal structure of lactobacillar SpaC reveals an atypical five-domain pilus tip adhesin: Exposing its substrate-binding and assembly in SpaCBA pili.
J.Struct.Biol., 211, 2020
|
|
8KB2
 
 | | Crystal Structure of M- and C-Domains of the shaft pilin LrpA from Ligilactobacillus ruminis - iodide derivative | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Prajapati, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2023-08-03 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The crystal structure of the N-terminal domain of the backbone pilin LrpA reveals a new closure-and-twist motion for assembling dynamic pili in Ligilactobacillus ruminis.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8KG4
 
 | | Crystal Structure of M- and C-Domains of the shaft pilin LrpA from Ligilactobacillus ruminis - orthorhombic form | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, IODIDE ION, ... | | Authors: | Prajapati, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2023-08-17 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of the N-terminal domain of the backbone pilin LrpA reveals a new closure-and-twist motion for assembling dynamic pili in Ligilactobacillus ruminis.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8KCL
 
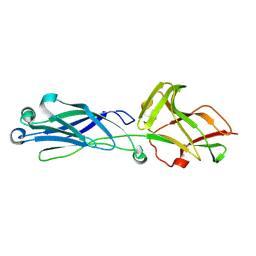 | |
5YU5
 
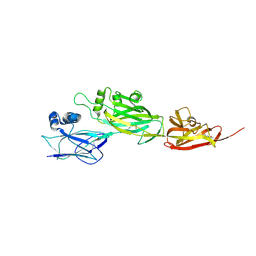 | | Crystal structure of shaft pilin spaD from Lactobacillus rhamnosus GG | | Descriptor: | Pilus assembly protein | | Authors: | Chaurasia, P, Pratap, S, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2017-11-20 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Bent conformation of a backbone pilin N-terminal domain supports a three-stage pilus assembly mechanism.
Commun Biol, 1, 2018
|
|
5YXO
 
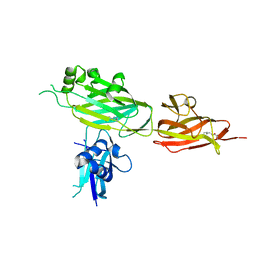 | | Crystal structure of shaft pilin spaD from Lactobacillus rhamnosus GG in bent conformation | | Descriptor: | Pilus assembly protein | | Authors: | Chaurasia, P, Pratap, S, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2017-12-06 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Bent conformation of a backbone pilin N-terminal domain supports a three-stage pilus assembly mechanism.
Commun Biol, 1, 2018
|
|
5YXG
 
 | | Crystal structure of C-terminal fragment of SpaD from Lactobacillus rhamnosus GG generated by limited proteolysis | | Descriptor: | CHLORIDE ION, Pilus assembly protein | | Authors: | Chaurasia, P, Pratap, S, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2017-12-05 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Bent conformation of a backbone pilin N-terminal domain supports a three-stage pilus assembly mechanism.
Commun Biol, 1, 2018
|
|
5Z0Z
 
 | | Crystal structure of shaft pilin spaD from Lactobacillus rhamnosus GG - D242A mutant | | Descriptor: | Pilus assembly protein | | Authors: | Chaurasia, P, Pratap, S, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2017-12-22 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Bent conformation of a backbone pilin N-terminal domain supports a three-stage pilus assembly mechanism.
Commun Biol, 1, 2018
|
|
5Z24
 
 | | Crystal structure of shaft pilin spaD from Lactobacillus rhamnosus GG - K365A mutant | | Descriptor: | Pilus assembly protein | | Authors: | Chaurasia, P, Pratap, S, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2017-12-28 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bent conformation of a backbone pilin N-terminal domain supports a three-stage pilus assembly mechanism.
Commun Biol, 1, 2018
|
|
6JCH
 
 | | Crystal structure of SpaE basal pilin from Lactobacillus rhamnosus GG - Orthorhombic form | | Descriptor: | Pilus assembly protein, SODIUM ION | | Authors: | Megta, A.K, Mishra, A.K, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2019-01-28 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.536 Å) | | Cite: | Crystal structure of basal pilin SpaE reveals the molecular basis of its incorporation in the lactobacillar SpaFED pilus.
J.Struct.Biol., 207, 2019
|
|
6JK7
 
 | | Crystal structure of SpaE basal pilin from Lactobacillus rhamnosus GG - Trigonal form | | Descriptor: | Pilus assembly protein | | Authors: | Megta, A.K, Mishra, A.K, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Crystal structure of basal pilin SpaE reveals the molecular basis of its incorporation in the lactobacillar SpaFED pilus.
J.Struct.Biol., 207, 2019
|
|
6JBV
 
 | | Crystal structure of SpaE basal pilin from Lactobacillus rhamnosus GG - Selenium derivative | | Descriptor: | Pilus assembly protein, SODIUM ION | | Authors: | Megta, A.K, Mishra, A.K, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2019-01-26 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Crystal structure of basal pilin SpaE reveals the molecular basis of its incorporation in the lactobacillar SpaFED pilus.
J.Struct.Biol., 207, 2019
|
|
5HTS
 
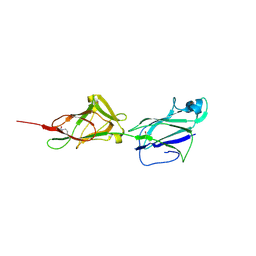 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG - D295N mutant | | Descriptor: | Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2016-01-27 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5HBB
 
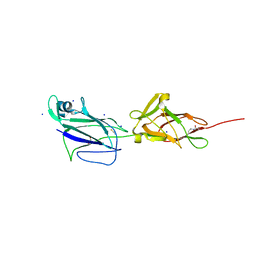 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG - E139A mutant | | Descriptor: | 1,2-ETHANEDIOL, Cell surface protein SpaA, SODIUM ION, ... | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-31 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5HDL
 
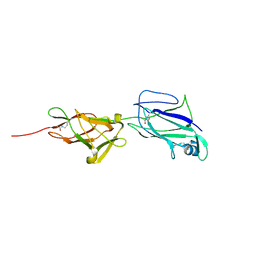 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG - E269A mutant | | Descriptor: | Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2016-01-05 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5J4M
 
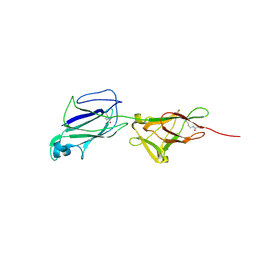 | | Crystal structure of shaft pilin SpaA from Lactobacillus rhamnosus GG - E269A/D295N double mutant | | Descriptor: | Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2016-04-01 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
1OC6
 
 | | structure native of the D405N mutant of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS at 1.5 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CELLOBIOHYDROLASE II, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-06 | | Release date: | 2003-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
