2E9S
 
 | | human neuronal Rab6B in three intermediate forms | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Vellieux, F.M, Tcherniuk, S, Garcia-Saez, I, Kozielski, F. | | Deposit date: | 2007-01-26 | | Release date: | 2008-01-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 3D structure of human neuronal Rab6B in three intermediate forms
To be Published
|
|
2WZN
 
 | | 3d structure of TET3 from Pyrococcus horikoshii | | Descriptor: | 354AA LONG HYPOTHETICAL OPERON PROTEIN FRV, CHLORIDE ION, GLYCEROL, ... | | Authors: | Rosenbaum, E, Dura, M.A, Vellieux, F.M, Franzetti, B. | | Deposit date: | 2009-12-01 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural and Biochemical Characterizations of a Novel Tet Peptidase Complex from Pyrococcus Horikoshii Reveal an Integrated Peptide Degradation System in Hyperthermophilic Archaea.
Mol.Microbiol., 72, 2009
|
|
4JCO
 
 | |
4JMQ
 
 | | Crystal structure of pb9: The Dit of bacteriophage T5. | | Descriptor: | Bacteriophage T5 distal tail protein | | Authors: | Flayhan, A, Vellieux, F.M.D, Girard, E, Maury, O, Boulanger, P, Breyton, C. | | Deposit date: | 2013-03-14 | | Release date: | 2013-11-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Crystal Structure of pb9, the Distal Tail Protein of Bacteriophage T5: a Conserved Structural Motif among All Siphophages.
J.Virol., 88, 2014
|
|
1BTM
 
 | | TRIOSEPHOSPHATE ISOMERASE (TIM) COMPLEXED WITH 2-PHOSPHOGLYCOLIC ACID | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Delboni, L.F, Mande, S.C, Hol, W.G.J. | | Deposit date: | 1995-11-11 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of recombinant triosephosphate isomerase from Bacillus stearothermophilus. An analysis of potential thermostability factors in six isomerases with known three-dimensional structures points to the importance of hydrophobic interactions.
Protein Sci., 4, 1995
|
|
1MDA
 
 | | CRYSTAL STRUCTURE OF AN ELECTRON-TRANSFER COMPLEX BETWEEN METHYLAMINE DEHYDROGENASE AND AMICYANIN | | Descriptor: | AMICYANIN, COPPER (II) ION, METHYLAMINE DEHYDROGENASE (HEAVY SUBUNIT), ... | | Authors: | Chen, L, Durley, R, Mathews, F.S. | | Deposit date: | 1992-03-02 | | Release date: | 1993-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an electron-transfer complex between methylamine dehydrogenase and amicyanin.
Biochemistry, 31, 1992
|
|
2J5R
 
 | | 2.25 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui after second radiation burn (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-19 | | Release date: | 2006-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
2J5Q
 
 | | 2.15 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui after first radiation burn (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-19 | | Release date: | 2006-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
2J5K
 
 | | 2.0 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-18 | | Release date: | 2006-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
1QUE
 
 | | X-RAY STRUCTURE OF THE FERREDOXIN:NADP+ REDUCTASE FROM THE CYANOBACTERIUM ANABAENA PCC 7119 AT 1.8 ANGSTROMS | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Serre, L, Frey, M, Vellieux, F.M.D. | | Deposit date: | 1996-07-06 | | Release date: | 1997-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of the ferredoxin:NADP+ reductase from the cyanobacterium Anabaena PCC 7119 at 1.8 A resolution, and crystallographic studies of NADP+ binding at 2.25 A resolution.
J.Mol.Biol., 263, 1996
|
|
1QUF
 
 | | X-RAY STRUCTURE OF A COMPLEX NADP+-FERREDOXIN:NADP+ REDUCTASE FROM THE CYANOBACTERIUM ANABAENA PCC 7119 AT 2.25 ANGSTROMS | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Serre, L, Frey, M, Vellieux, F.M.D. | | Deposit date: | 1996-09-07 | | Release date: | 1997-09-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray structure of the ferredoxin:NADP+ reductase from the cyanobacterium Anabaena PCC 7119 at 1.8 A resolution, and crystallographic studies of NADP+ binding at 2.25 A resolution.
J.Mol.Biol., 263, 1996
|
|
2X0J
 
 | | 2.8 A RESOLUTION STRUCTURE OF MALATE DEHYDROGENASE FROM ARCHAEOGLOBUS FULGIDUS IN COMPLEX WITH ETHENO-NAD | | Descriptor: | ETHENO-NAD, MALATE DEHYDROGENASE, SULFATE ION | | Authors: | Irimia, A, Madern, D, Zaccai, G, Vellieux, F.M, Karshikoff, A, Tibbelin, G, Ladenstein, R, Lien, T, Birkeland, N.K. | | Deposit date: | 2009-12-14 | | Release date: | 2009-12-22 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (2.786 Å) | | Cite: | The 2.9A Resolution Crystal Structure of Malate Dehydrogenase from Archaeoglobus Fulgidus: Mechanisms of Oligomerisation and Thermal Stabilisation.
J.Mol.Biol., 335, 2004
|
|
1W35
 
 | | FERREDOXIN-NADP+ REDUCTASE (MUTATION: Y 303 W) | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Perez-Dorado, I, Medina, M, Julvez, M.M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2004-07-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | C-Terminal Tyrosine of Ferredoxin-Nadp(+) Reductase in Hydride Transfer Processes with Nad(P)(+)/H.
Biochemistry, 44, 2005
|
|
1B2R
 
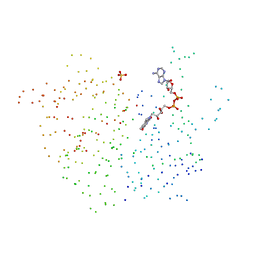 | | FERREDOXIN-NADP+ REDUCTASE (MUTATION: E 301 A) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN-NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1998-11-27 | | Release date: | 1999-12-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the catalytic role of Glu301 in Anabaena PCC 7119 ferredoxin-NADP+ reductase revealed by x-ray crystallography.
Proteins, 38, 2000
|
|
1BJK
 
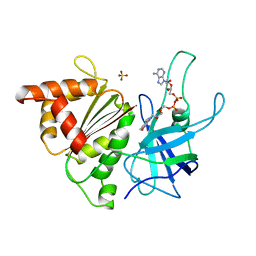 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH ARG 264 REPLACED BY GLU (R264E) | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1998-06-25 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Arg100 and Arg264 from Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal NADP+ binding and electron transfer.
Biochemistry, 37, 1998
|
|
1BQE
 
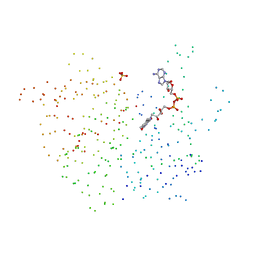 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH THR 155 REPLACED BY GLY (T155G) | | Descriptor: | FERREDOXIN--NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1998-08-14 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Probing the determinants of coenzyme specificity in ferredoxin-NADP+ reductase by site-directed mutagenesis.
J.Biol.Chem., 276, 2001
|
|
5MF2
 
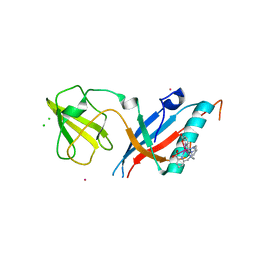 | | Bacteriophage T5 distal tail protein pb9 co-crystallized with Tb-Xo4 | | Descriptor: | CHLORIDE ION, Distal tail protein, TERBIUM(III) ION, ... | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Lassalle, L, Arnaud, C.-A, Breyton, C, Madern, D, Coquelle, N, Maury, O, Girard, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-09-20 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallophore: a versatile lanthanide complex for protein crystallography combining nucleating effects, phasing properties, and luminescence.
Chem Sci, 8, 2017
|
|
1QGZ
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LEU 78 REPLACED BY ASP (L78D) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1999-05-10 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of a cluster of hydrophobic residues near the FAD cofactor in Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal complex formation and electron transfer to ferredoxin.
J.Biol.Chem., 276, 2001
|
|
1QH0
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LEU 76 MUTATED BY ASP AND LEU 78 MUTATED BY ASP | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1999-05-10 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Role of a cluster of hydrophobic residues near the FAD cofactor in Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal complex formation and electron transfer to ferredoxin.
J.Biol.Chem., 276, 2001
|
|
1QGY
 
 | | Ferredoxin:NADP+ reductase mutant with Lys 75 replaced by Glu (K75E) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP+ reductase, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1999-05-10 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of interactions for complex formation between Ferredoxin-NADP+ reductase and its protein partners
Proteins, 59, 2005
|
|
