1G41
 
 | | CRYSTAL STRUCTURE OF HSLU HAEMOPHILUS INFLUENZAE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK PROTEIN HSLU, SULFATE ION | | Authors: | Trame, C.B, McKay, D.B. | | Deposit date: | 2000-10-25 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Haemophilus influenzae HslU protein in crystals with one-dimensional disorder twinning.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1N5B
 
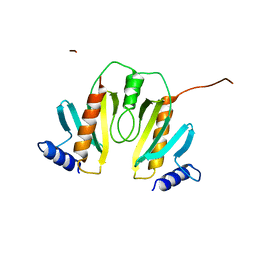 | |
1IM2
 
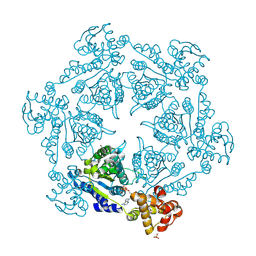 | | HslU, Haemophilus Influenzae, Selenomethionine Variant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, SULFATE ION | | Authors: | Trame, C.B, McKay, D.B. | | Deposit date: | 2001-05-09 | | Release date: | 2001-08-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Haemophilus influenzae HslU protein in crystals with one-dimensional disorder twinning.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G3I
 
 | | CRYSTAL STRUCTURE OF THE HSLUV PROTEASE-CHAPERONE COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-DEPENDENT HSLU PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Sousa, M.C, Trame, C.B, Tsuruta, H, Wilbanks, S.M, Reddy, V.S, McKay, D.B. | | Deposit date: | 2000-10-24 | | Release date: | 2000-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Crystal and solution structures of an HslUV protease-chaperone complex.
Cell(Cambridge,Mass.), 103, 2000
|
|
1IKP
 
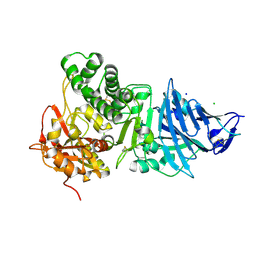 | | Pseudomonas Aeruginosa Exotoxin A, P201Q, W281A mutant | | Descriptor: | CHLORIDE ION, EXOTOXIN A, SODIUM ION | | Authors: | McKay, D.B, Wedekind, J.E, Trame, C.B. | | Deposit date: | 2001-05-04 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Refined Crystallographic Structure of Pseudomonas aeruginosa Exotoxin A
and its Implications for the Molecular Mechanism of Toxicity
J.Mol.Biol., 314, 2001
|
|
1IKQ
 
 | | Pseudomonas Aeruginosa Exotoxin A, wild type | | Descriptor: | CHLORIDE ION, EXOTOXIN A, SODIUM ION | | Authors: | McKay, D.B, Wedekind, J.E, Trame, C.B. | | Deposit date: | 2001-05-04 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Refined Crystallographic Structure of Pseudomonas aeruginosa Exotoxin A
and its Implications for the Molecular Mechanism of Toxicity
J.Mol.Biol., 314, 2001
|
|
3NL9
 
 | |
2GLZ
 
 | |
2GVI
 
 | |
1G3K
 
 | |
3GF8
 
 | |
3H41
 
 | |
3H0N
 
 | |
3HBZ
 
 | |
3HSA
 
 | |
3K5J
 
 | |
3KK7
 
 | |
1KYI
 
 | | HslUV (H. influenzae)-NLVS Vinyl Sulfone Inhibitor Complex | | Descriptor: | 4-IODO-3-NITROPHENYL ACETYL-LEUCINYL-LEUCINYL-LEUCINYL-VINYLSULFONE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent hsl protease ATP-binding subunit hslU, ... | | Authors: | Sousa, M.C, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | Deposit date: | 2002-02-04 | | Release date: | 2002-05-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of HslUV Complexed with a Vinyl Sulfone Inhibitor:
Corroboration of a Proposed Mechanism of Allosteric Activation of
HslV by HslU
J.Mol.Biol., 318, 2002
|
|
3B77
 
 | |
3BYQ
 
 | |
3D00
 
 | |
3CGH
 
 | |
3CM1
 
 | |
3DEE
 
 | |
3DCX
 
 | |
