1Z17
 
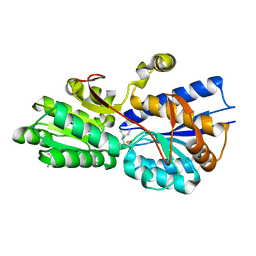 | | Crystal structure analysis of periplasmic Leu/Ile/Val-binding protein with bound ligand isoleucine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ISOLEUCINE, Leu/Ile/Val-binding protein | | Authors: | Trakhanov, S.D, Vyas, N.K, Kristensen, D.M, Ma, J, Quiocho, F.A. | | Deposit date: | 2005-03-03 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity.
Biochemistry, 44, 2005
|
|
1Z18
 
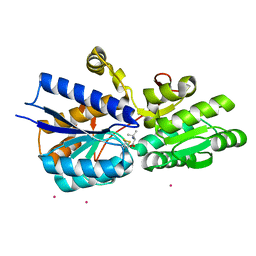 | | Crystal structure analysis of periplasmic Leu/Ile/Val-binding protein with bound valine | | Descriptor: | CADMIUM ION, Leu/Ile/Val-binding protein, VALINE | | Authors: | Trakhanov, S.D, Vyas, N.K, Kristensen, D.M, Ma, J, Quiocho, F.A. | | Deposit date: | 2005-03-03 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity.
Biochemistry, 44, 2005
|
|
1Z16
 
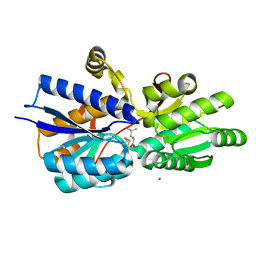 | | Crystal structure analysis of periplasmic Leu/Ile/Val-binding protein with bound leucine | | Descriptor: | CADMIUM ION, LEUCINE, Leu/Ile/Val-binding protein | | Authors: | Trakhanov, S.D, Vyas, N.K, Kristensen, D.M, Ma, J, Quiocho, F.A. | | Deposit date: | 2005-03-03 | | Release date: | 2005-10-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity.
Biochemistry, 44, 2005
|
|
1Z15
 
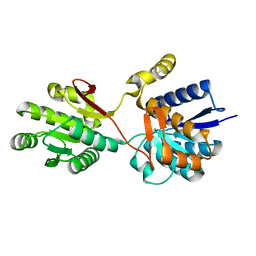 | | Crystal structure analysis of periplasmic Leu/Ile/Val-binding protein in superopen form | | Descriptor: | Leu/Ile/Val-binding protein | | Authors: | Trakhanov, S.D, Vyas, N.K, Kristensen, D.M, Ma, J, Quiocho, F.A. | | Deposit date: | 2005-03-03 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity.
Biochemistry, 44, 2005
|
|
2LBP
 
 | |
1ACX
 
 | |
1HSL
 
 | |
1BZ4
 
 | | APOLIPOPROTEIN E3 (APO-E3), TRUNCATION MUTANT 165 | | Descriptor: | PROTEIN (APOLIPOPROTEIN E) | | Authors: | Rupp, B, Segelke, B. | | Deposit date: | 1998-11-05 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational flexibility in the apolipoprotein E amino-terminal domain structure determined from three new crystal forms: implications for lipid binding.
Protein Sci., 9, 2000
|
|
1NTN
 
 | | THE CRYSTAL STRUCTURE OF NEUROTOXIN-I FROM NAJA NAJA OXIANA AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | NEUROTOXIN I | | Authors: | Mikhailov, A.M, Nickitenko, A.V, Vainshtein, B.K, Betzel, C, Wilson, K. | | Deposit date: | 1994-09-26 | | Release date: | 1995-05-08 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-dimensional structure of neurotoxin-1 from Naja naja oxiana venom at 1.9 A resolution.
Febs Lett., 320, 1993
|
|
1NFN
 
 | | APOLIPOPROTEIN E3 (APOE3) | | Descriptor: | APOLIPOPROTEIN E3 | | Authors: | Rupp, B, Parkin, S. | | Deposit date: | 1996-07-17 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia.
Nat.Struct.Biol., 3, 1996
|
|
1NFO
 
 | | APOLIPOPROTEIN E2 (APOE2, D154A MUTATION) | | Descriptor: | APOLIPOPROTEIN E2 | | Authors: | Rupp, B, Parkin, S. | | Deposit date: | 1996-07-17 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia.
Nat.Struct.Biol., 3, 1996
|
|
1OR3
 
 | | APOLIPOPROTEIN E3 (APOE3), TRIGONAL TRUNCATION MUTANT 165 | | Descriptor: | PROTEIN (APOLIPOPROTEIN E) | | Authors: | Rupp, B, Segelke, B.W. | | Deposit date: | 1998-12-01 | | Release date: | 2000-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Conformational flexibility in the apolipoprotein E amino-terminal domain structure determined from three new crystal forms: implications for lipid binding.
Protein Sci., 9, 2000
|
|
1OR2
 
 | | APOLIPOPROTEIN E3 (APOE3) TRUNCATION MUTANT 165 | | Descriptor: | APOLIPOPROTEIN E | | Authors: | Rupp, B, Segelke, B.W, Forstner, M. | | Deposit date: | 1999-03-25 | | Release date: | 2000-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational flexibility in the apolipoprotein E amino-terminal domain structure determined from three new crystal forms: implications for lipid binding.
Protein Sci., 9, 2000
|
|
2LIV
 
 | |
