1AMX
 
 | |
1YIS
 
 | | Structural genomics of Caenorhabditis elegans: adenylosuccinate lyase | | Descriptor: | SULFATE ION, adenylosuccinate lyase | | Authors: | Symersky, J, Schormann, N, Lu, S, Zhang, Y, Karpova, E, Qiu, S, Huang, W, Cao, Z, Zhou, J, Luo, M, Arabshahi, A, McKinstry, A, Luan, C.-H, Luo, D, Johnson, D, An, J, Tsao, J, Delucas, L, Shang, Q, Gray, R, Li, S, Bray, T, Chen, Y.-J, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-01-12 | | Release date: | 2005-01-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: adenylosuccinate lyase
To be Published
|
|
2EUL
 
 | | Structure of the transcription factor Gfh1. | | Descriptor: | ZINC ION, anti-cleavage anti-GreA transcription factor Gfh1 | | Authors: | Symersky, J, Perederina, A, Vassylyeva, M.N, Svetlov, V, Artsimovitch, I, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-28 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulation through the RNA Polymerase Secondary Channel: STRUCTURAL AND FUNCTIONAL VARIABILITY OF THE COILED-COIL TRANSCRIPTION FACTORS.
J.Biol.Chem., 281, 2006
|
|
1KQP
 
 | | NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS AT 1 A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, ... | | Authors: | Symersky, J, Devedjiev, Y, Moore, K, Brouillette, C, DeLucas, L. | | Deposit date: | 2002-01-07 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | NH3-dependent NAD+ synthetase from Bacillus subtilis at 1 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1T9F
 
 | | Structural genomics of Caenorhabditis elegans: Structure of a protein with unknown function | | Descriptor: | MALONATE ION, protein 1d10 | | Authors: | Symersky, J, Li, S, Bunzel, R, Schormann, N, Luo, D, Huang, W.Y, Qiu, S, Gray, R, Zhang, Y, Arabashi, A, Lu, S, Luan, C.H, Tsao, J, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-16 | | Release date: | 2004-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: Structure of a protein with unknown function.
To be Published
|
|
1MO0
 
 | | Structural Genomics Of Caenorhabditis Elegans: Triose Phosphate Isomerase | | Descriptor: | ACETATE ION, SULFATE ION, Triosephosphate isomerase | | Authors: | Symersky, J, Li, S, Finley, J, Liu, Z.-J, Qui, H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, DeLucas, L.J, Wang, B.-C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2002-09-06 | | Release date: | 2002-09-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: triosephosphate isomerase
Proteins, 51, 2003
|
|
3U2Y
 
 | | ATP synthase c10 ring in proton-unlocked conformation at pH 6.1 | | Descriptor: | ATP synthase subunit C, mitochondrial | | Authors: | Symersky, J, Pagadala, V, Osowski, D, Krah, A, Meier, T, Faraldo-Gomez, J, Mueller, D.M. | | Deposit date: | 2011-10-04 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the c(10) ring of the yeast mitochondrial ATP synthase in the open conformation.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3U32
 
 | | ATP synthase c10 ring reacted with DCCD at pH 5.5 | | Descriptor: | ATP synthase subunit C, mitochondrial, DICYCLOHEXYLUREA | | Authors: | Symersky, J, Pagadala, V, Osowski, D, Krah, A, Meier, T, Faraldo-Gomez, J, Mueller, D.M. | | Deposit date: | 2011-10-04 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the c(10) ring of the yeast mitochondrial ATP synthase in the open conformation.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3U2F
 
 | | ATP synthase c10 ring in proton-unlocked conformation at PH 8.3 | | Descriptor: | ATP synthase subunit C, mitochondrial | | Authors: | Symersky, J, Pagadala, V, Osowski, D, Krah, A, Meier, T, Faraldo-Gomez, J, Mueller, D.M. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the c(10) ring of the yeast mitochondrial ATP synthase in the open conformation.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UD0
 
 | |
1T7S
 
 | | Structural Genomics of Caenorhabditis elegans: Structure of BAG-1 protein | | Descriptor: | BAG-1 cochaperone | | Authors: | Symersky, J, Zhang, Y, Schormann, N, Li, S, Bunzel, R, Pruett, P, Luan, C.-H, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-10 | | Release date: | 2004-05-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: structure of the BAG domain.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4F4S
 
 | |
1J71
 
 | |
1OOJ
 
 | | Structural genomics of Caenorhabditis elegans : Calmodulin | | Descriptor: | CALCIUM ION, Calmodulin CMD-1 | | Authors: | Symersky, J, Lin, G, Li, S, Qiu, S, Luan, C.-H, Luo, D, Tsao, J, Carson, M, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-03-03 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural genomics of caenorhabditis elegans: crystal structure of calmodulin.
Proteins, 53, 2003
|
|
1OOE
 
 | | Structural Genomics of Caenorhabditis elegans : Dihydropteridine reductase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dihydropteridine reductase | | Authors: | Symersky, J, Li, S, Nagy, L, Qiu, S, Lin, G, Tsao, J, Luo, D, Carson, M, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-03-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: structure of dihydropteridine reductase.
Proteins, 53, 2003
|
|
3CLF
 
 | |
1PGV
 
 | | Structural Genomics of Caenorhabditis elegans: tropomodulin C-terminal domain | | Descriptor: | tropomodulin TMD-1 | | Authors: | Symersky, J, Lu, S, Li, S, Chen, L, Meehan, E, Luo, M, Qiu, S, Bunzel, R.J, Luo, D, Arabashi, A, Nagy, L.A, Lin, G, Luan, W.C.-H, Carson, M, Gray, R, Huang, W, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-05-28 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: crystal structure of the tropomodulin C-terminal domain
Proteins, 56, 2004
|
|
5BQ6
 
 | |
5BQJ
 
 | |
5BQA
 
 | |
5BPS
 
 | |
2B34
 
 | | Structure of MAR1 Ribonuclease from Caenorhabditis elegans | | Descriptor: | MAR1 Ribonuclease | | Authors: | Schormann, N, Karpova, E, Li, S, Symersky, J, Zhang, Y, Lu, S, Zhou, Q, Lin, G, Cao, Z, Luo, M, Qiu, S, Luan, C.-H, Luo, D, Huang, W, Shang, Q, McKinstry, A, An, J, Tsao, J, Carson, M, Stinnett, M, Chen, Y, Johnson, D, Gary, R, Arabshahi, A, Bunzel, R, Bray, T, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-09-19 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structure of MAR1 Ribonuclease from Caenorhabditis elegans
To be Published
|
|
2GP4
 
 | |
2PNQ
 
 | |
6CP5
 
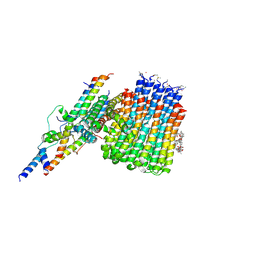 | | Monomer yeast ATP synthase Fo reconstituted in nanodisc with inhibitor of oligomycin bound generated from focused refinement. | | Descriptor: | ATP synthase protein 8, ATP synthase subunit 4, mitochondrial, ... | | Authors: | Srivastava, A.P, Luo, M, Symersky, J, Liao, M.F, Mueller, D.M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-04-11 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane.
Science, 360, 2018
|
|
