3IDQ
 
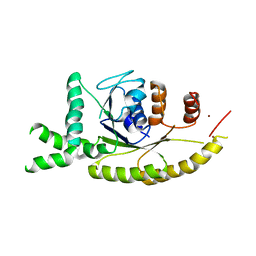 | | Crystal structure of S. cerevisiae Get3 at 3.7 Angstrom resolution | | Descriptor: | ATPase GET3, NICKEL (II) ION, ZINC ION | | Authors: | Suloway, C.J.M, Chartron, J.W, Zaslaver, M, Clemons Jr, W.M. | | Deposit date: | 2009-07-21 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.701 Å) | | Cite: | Model for eukaryotic tail-anchored protein binding based on the structure of Get3
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3UG7
 
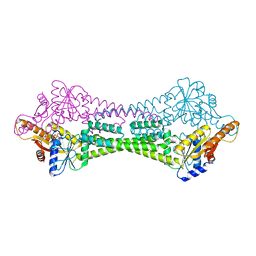 | | Crystal Structure of Get3 from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Suloway, C.J.M, Rome, M.E, Clemons Jr, W.M. | | Deposit date: | 2011-11-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Tail-anchor targeting by a Get3 tetramer: the structure of an archaeal homologue.
Embo J., 31, 2012
|
|
3IBG
 
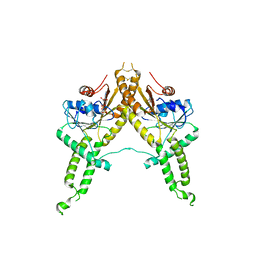 | | Crystal structure of Aspergillus fumigatus Get3 with bound ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase, subunit of the Get complex | | Authors: | Suloway, C.J.M, Chartron, J.W, Zaslaver, M, Clemons Jr, W.M. | | Deposit date: | 2009-07-15 | | Release date: | 2009-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Model for eukaryotic tail-anchored protein binding based on the structure of Get3
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3UG6
 
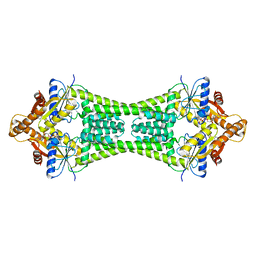 | | Crystal Structure of Get3 from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Suloway, C.J.M, Rome, M.E, Clemons Jr, W.M. | | Deposit date: | 2011-11-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Tail-anchor targeting by a Get3 tetramer: the structure of an archaeal homologue.
Embo J., 31, 2012
|
|
4HTT
 
 | |
