4L6Y
 
 | | Structure of the microtubule associated protein PRC1 (Protein Regulator of Cytokinesis 1) | | Descriptor: | Protein regulator of cytokinesis 1 | | Authors: | Subramanian, R, Ti, S, Tan, L, Darst, S.A, Kapoor, T.M. | | Deposit date: | 2013-06-13 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3015 Å) | | Cite: | Marking and Measuring Single Microtubules by PRC1 and Kinesin-4.
Cell(Cambridge,Mass.), 154, 2013
|
|
4L3I
 
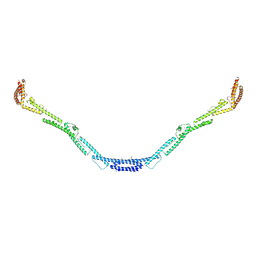 | | Structure of the microtubule associated protein PRC1 (Protein Regulator of Cytokinesis 1) | | Descriptor: | Protein regulator of cytokinesis 1 | | Authors: | Subramanian, R, Ti, S, Tan, L, Darst, S.A, Kapoor, T.M. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.6005 Å) | | Cite: | Marking and Measuring Single Microtubules by PRC1 and Kinesin-4.
Cell(Cambridge,Mass.), 154, 2013
|
|
8F0V
 
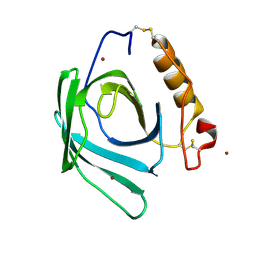 | | Lipocalin-like Milk protein-2 - E38A mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Milk protein, ZINC ION | | Authors: | Subramanian, R, KanagaVijayan, D. | | Deposit date: | 2022-11-04 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Variability in phenylalanine side chain conformations facilitates broad substrate tolerance of fatty acid binding in cockroach milk proteins.
Plos One, 18, 2023
|
|
8F0Y
 
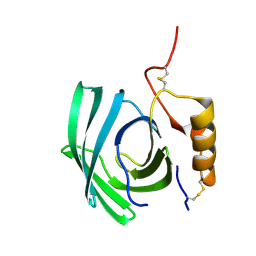 | | Lipocalin-like Milk protein-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Milk protein | | Authors: | Subramanian, R, KanagaVijayan, D, Shantakumar, R.P.S. | | Deposit date: | 2022-11-04 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Variability in phenylalanine side chain conformations facilitates broad substrate tolerance of fatty acid binding in cockroach milk proteins.
Plos One, 18, 2023
|
|
7YX1
 
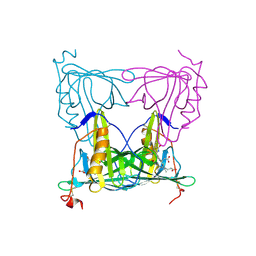 | |
3NRX
 
 | | Insights into anti-parallel microtubule crosslinking by PRC1, a conserved non-motor microtubule binding protein | | Descriptor: | Protein regulator of cytokinesis 1 | | Authors: | Kapoor, T.M, Subramanian, R, Wilson-Kubalek, E.M, Arthur, C.P, Bick, M.J, Campbell, E.A, Darst, S.A, Milligan, R.A. | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into Antiparallel Microtubule Crosslinking by PRC1, a Conserved Nonmotor Microtubule Binding Protein.
Cell(Cambridge,Mass.), 142, 2010
|
|
3NRY
 
 | | Insights into anti-parallel microtubule crosslinking by PRC1, a conserved microtubule binding protein | | Descriptor: | Protein regulator of cytokinesis 1 | | Authors: | Kapoor, T.M, Subramanian, R, Wilson-Kubalek, E.M, Arthur, C.P, Bick, M.J, Campbell, E.A, Darst, S.A, Milligan, R.A. | | Deposit date: | 2010-07-01 | | Release date: | 2010-08-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into Antiparallel Microtubule Crosslinking by PRC1, a Conserved Nonmotor Microtubule Binding Protein.
Cell(Cambridge,Mass.), 142, 2010
|
|
7LQM
 
 | |
7LQN
 
 | |
7KCB
 
 | | Symmetry in Yeast Alcohol Dehydrogenase 1 -Closed Form with NAD+ and Trifluoroethanol | | Descriptor: | ADH1 isoform 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL, ... | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V, Guntupalli, S.R. | | Deposit date: | 2020-10-05 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
7KCQ
 
 | | Symmetry in Yeast Alcohol Dehydrogenase 1 -Open Form of Apoenzyme | | Descriptor: | Alcohol dehydrogenase, ZINC ION | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V, Guntupalli, S.R. | | Deposit date: | 2020-10-07 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
7KJY
 
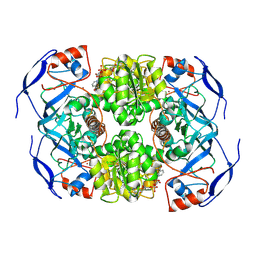 | | Symmetry in Yeast Alcohol Dehydrogenase 1 - Open Form with NADH | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V, Guntupalli, S.R. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
7KC2
 
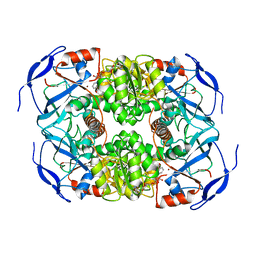 | | Symmetry in Yeast Alcohol Dehydrogenase 1 -Closed Form with NADH | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V. | | Deposit date: | 2020-10-04 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
5YYB
 
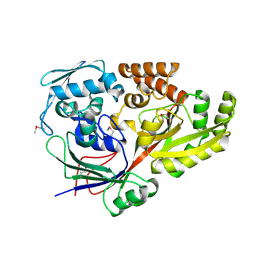 | |
5Z99
 
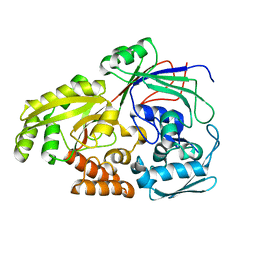 | |
6LAY
 
 | |
6LVE
 
 | | Structure of Dimethylformamidase, tetramer, E521A mutant | | Descriptor: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LVC
 
 | | Structure of Dimethylformamidase, dimer | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7BKX
 
 | | Diploptera punctata inspired lipocalin-like Milk protein expressed in Saccharomyces cerevisiae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Banerjee, S, Kanagavijayan, D, Subramanian, R, Santhakumari, P.R, Chavas, L.M.G, Ramaswamy, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of recombinantly expressed cockroach Lili-Mip protein in glycosylated and deglycosylated forms.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
6IFD
 
 | |
6JOV
 
 | |
6IFI
 
 | |
6LVB
 
 | | Structure of Dimethylformamidase, tetramer | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
9BPC
 
 | | Cryo-EM structure of P2X3 receptor in complex with camlipixant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Camlipixant, ... | | Authors: | Thach, T, Subramanian, R. | | Deposit date: | 2024-05-07 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Mechanistic insights into the selective targeting of P2X3 receptor by camlipixant antagonist.
J.Biol.Chem., 301, 2025
|
|
9BPD
 
 | | Cryo-EM structure of P2X3 receptor in complex with ATP:Mg2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Thach, T, Subramanian, R. | | Deposit date: | 2024-05-07 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Mechanistic insights into the selective targeting of P2X3 receptor by camlipixant antagonist.
J.Biol.Chem., 301, 2025
|
|
