1BEV
 
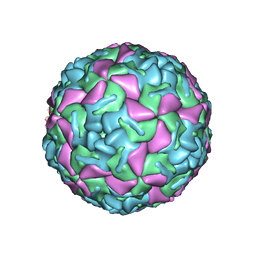 | | BOVINE ENTEROVIRUS VG-5-27 | | Descriptor: | BOVINE ENTEROVIRUS COAT PROTEINS VP1 TO VP4, MYRISTIC ACID, SULFATE ION | | Authors: | Smyth, M, Tate, J, Lyons, C, Hoey, E, Martin, S, Stuart, D. | | Deposit date: | 1996-04-03 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Implications for viral uncoating from the structure of bovine enterovirus.
Nat.Struct.Biol., 2, 1995
|
|
2AIA
 
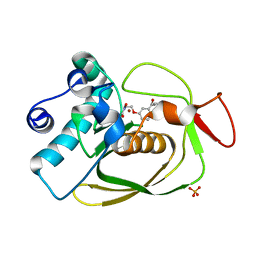 | | S.pneumoniae PDF complexed with SB-543668 | | Descriptor: | 2-(3-BENZOYLPHENOXY)ETHYL(HYDROXY)FORMAMIDE, NICKEL (II) ION, Peptide deformylase, ... | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species.
Protein Sci., 12, 2003
|
|
1D4M
 
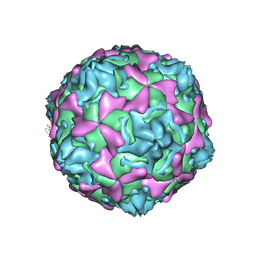 | | THE CRYSTAL STRUCTURE OF COXSACKIEVIRUS A9 TO 2.9 A RESOLUTION | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, MYRISTIC ACID, PROTEIN (COXSACKIEVIRUS A9) | | Authors: | Hendry, E, Hatanaka, H, Fry, E, Smyth, M, Tate, J, Stanway, G, Santti, J, Maaronen, M, Hyypia, T, Stuart, D. | | Deposit date: | 1999-10-04 | | Release date: | 1999-12-23 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses.
Structure Fold.Des., 7, 1999
|
|
2AIE
 
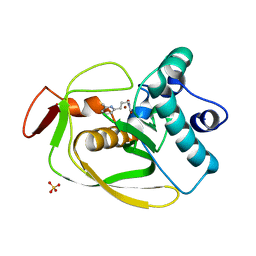 | | S.pneumoniae polypeptide deformylase complexed with SB-505684 | | Descriptor: | HYDROXY[3-(6-METHYLPYRIDIN-2-YL)PROPYL]FORMAMIDE, NICKEL (II) ION, Peptide deformylase, ... | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species
Protein Sci., 12, 2003
|
|
2AI8
 
 | | E.coli Polypeptide Deformylase complexed with SB-485343 | | Descriptor: | NICKEL (II) ION, Peptide deformylase, [HYDROXY(3-PHENYLPROPYL)AMINO]METHANOL | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species.
Protein Sci., 12, 2003
|
|
2AI7
 
 | | S.pneumoniae Polypeptide Deformylase complexed with SB-485345 | | Descriptor: | NICKEL (II) ION, Peptide deformylase, SULFATE ION, ... | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species
Protein Sci., 12, 2003
|
|
2AI9
 
 | | S.aureus Polypeptide Deformylase | | Descriptor: | NICKEL (II) ION, Peptide deformylase, SULFATE ION | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species.
Protein Sci., 12, 2003
|
|
1C14
 
 | | CRYSTAL STRUCTURE OF E COLI ENOYL REDUCTASE-NAD+-TRICLOSAN COMPLEX | | Descriptor: | ENOYL REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Qiu, X, Janson, C, Court, R, Smyth, M, Payne, D, Abdel-Meguid, S. | | Deposit date: | 1999-07-20 | | Release date: | 2000-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for triclosan activity involves a flipping loop in the active site.
Protein Sci., 8, 1999
|
|
3SW8
 
 | |
