2WZL
 
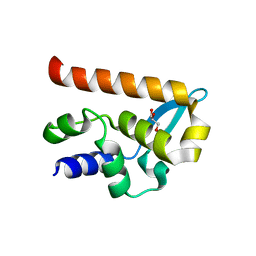 | | The Structure of the N-RNA Binding Domain of the Mokola virus Phosphoprotein | | Descriptor: | GLYCEROL, PHOSPHOPROTEIN | | Authors: | Assenberg, R, Delmas, O, Ren, J, Vidalain, P, Verma, A, Larrous, F, Graham, S, Tangy, F, Grimes, J, Bourhy, H. | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the N-RNA Binding Domain of the Mokola Virus Phosphoprotein
J.Virol., 84, 2010
|
|
3KI0
 
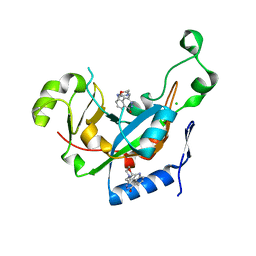 | | Catalytic fragment of Cholix toxin from Vibrio Cholerae in complex with inhibitor GP-D | | Descriptor: | 3-(morpholin-4-ylmethyl)-1,5-dihydro-6H-[1,2]diazepino[4,5,6-cd]indol-6-one, CHLORIDE ION, Cholix toxin | | Authors: | Jorgensen, R, Edwards, P.R, Merrill, A.R. | | Deposit date: | 2009-10-31 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structure function analysis of soluble inhibitors of cholix toxin from Vibrio cholerae
To be Published
|
|
3KI7
 
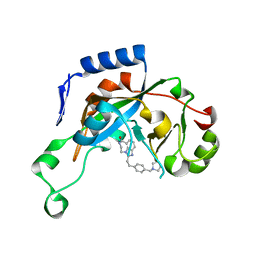 | | Catalytic fragment of Cholix toxin from Vibrio Cholerae in complex with inhibitor GP-I | | Descriptor: | 2-{2-[4-(pyrrolidin-1-ylmethyl)phenyl]ethyl}-5,6-dihydroimidazo[4,5,1-jk][1,4]benzodiazepin-7(4H)-one, Cholix toxin | | Authors: | Jorgensen, R, Edwards, P.R, Merrill, A.R. | | Deposit date: | 2009-10-31 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structure function analysis of soluble inhibitors of cholix toxin from Vibrio cholerae
To be Published
|
|
3KI2
 
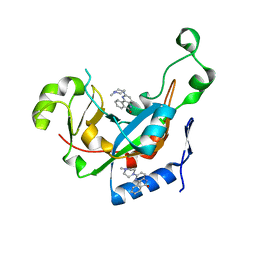 | |
3KI4
 
 | | Catalytic fragment of Cholix toxin from Vibrio Cholerae in complex with inhibitor GP-P | | Descriptor: | (11bR)-3-oxo-1,2,3,11b-tetrahydrochromeno[4,3,2-de]isoquinoline-10-sulfonic acid, Cholix toxin, GLYCEROL | | Authors: | Jorgensen, R, Edwards, P.R, Merrill, A.R. | | Deposit date: | 2009-10-31 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure function analysis of soluble inhibitors of cholix toxin from Vibrio cholerae
To be Published
|
|
3KI3
 
 | | Catalytic fragment of Cholix toxin from Vibrio Cholerae in complex with inhibitor GP-H | | Descriptor: | 2-[(dimethylamino)methyl]-5,6-dihydroimidazo[4,5,1-jk][1,4]benzodiazepin-7(4H)-one, Cholix toxin | | Authors: | Jorgensen, R, Edwards, P.R, Merrill, A.R. | | Deposit date: | 2009-10-31 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structure function analysis of soluble inhibitors of cholix toxin from Vibrio cholerae
To be Published
|
|
3KI6
 
 | | Catalytic fragment of Cholix toxin from Vibrio Cholerae in complex with inhibitor GP-L | | Descriptor: | 8-fluoro-2-(3-piperidin-1-ylpropanoyl)-1,3,4,5-tetrahydrobenzo[c][1,6]naphthyridin-6(2H)-one, Cholix toxin | | Authors: | Jorgensen, R, Edwards, P.R, Merrill, A.R. | | Deposit date: | 2009-10-31 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure function analysis of soluble inhibitors of cholix toxin from Vibrio cholerae
To be Published
|
|
3KI5
 
 | |
4X2E
 
 | | Clostridium difficile wild type Fic protein | | Descriptor: | Fic family protein putative filamentation induced by cAMP protein | | Authors: | Jorgensen, R, Dedic, E. | | Deposit date: | 2014-11-26 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Structure of wild type Clostridium difficile Fic_0569 at 3.1 Angstroms resolution
To Be Published
|
|
4X2C
 
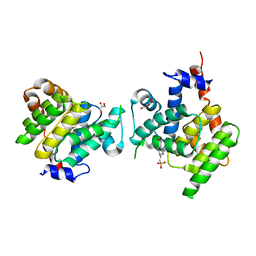 | | Clostridium difficile Fic protein_0569 mutant S31A, E35A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fic family protein putative filamentation induced by cAMP protein, GLYCEROL, ... | | Authors: | Jorgensen, R, Dedic, E. | | Deposit date: | 2014-11-26 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Clostridium difficile Fic_0569 S31A, E35A mutant at 1.8 Angstroms resolution
To Be Published
|
|
4X2D
 
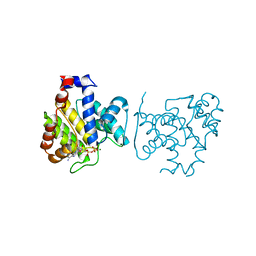 | |
3KI1
 
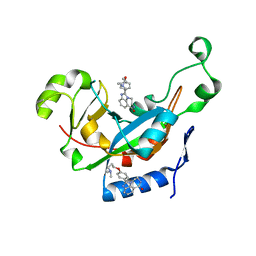 | | Catalytic fragment of Cholix toxin from Vibrio Cholerae in complex with inhibitor GP-F | | Descriptor: | 2-{4-[3-(dimethylamino)propoxy]phenyl}-5,6-dihydroimidazo[4,5,1-jk][1,4]benzodiazepin-7(4H)-one, Cholix toxin | | Authors: | Jorgensen, R, Edwards, P.R, Merrill, A.R. | | Deposit date: | 2009-10-31 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structure function analysis of soluble inhibitors of cholix toxin from Vibrio cholerae
To be Published
|
|
3LE4
 
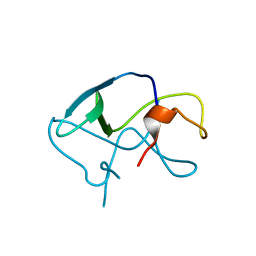 | |
6B0V
 
 | | Crystal Structure of small molecule ARS-107 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-(4-{[(4,5-dichloro-2-hydroxyphenyl)amino]acetyl}piperazin-1-yl)azetidin-1-yl]propan-1-one, CALCIUM ION, GTPase KRas, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6B0Y
 
 | | Crystal Structure of small molecule ARS-917 covalently bound to K-Ras G12C | | Descriptor: | 1-{4-[6-chloro-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5JWQ
 
 | | Crystal structure of KaiC S431E in complex with foldswitch-stabilized KaiB from Thermosynechococcus elongatus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiB, Circadian clock protein kinase KaiC | | Authors: | Tseng, R, Goularte, N.F, Chavan, A, Luu, J, Chang, Y, Heilser, J, Tripathi, S, LiWang, A, Partch, C.L. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.871 Å) | | Cite: | Structural basis of the day-night transition in a bacterial circadian clock.
Science, 355, 2017
|
|
5JWO
 
 | | Crystal structure of foldswitch-stabilized KaiB in complex with the N-terminal CI domain of KaiC from Thermosynechococcus elongatus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiB, Circadian clock protein kinase KaiC | | Authors: | Tseng, R, Goularte, N.F, Chavan, A, Luu, J, Chang, Y, Heilser, J, Tripathi, S, LiWang, A, Partch, C.L. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the day-night transition in a bacterial circadian clock.
Science, 355, 2017
|
|
5JWR
 
 | | Crystal structure of foldswitch-stabilized KaiB in complex with the N-terminal CI domain of KaiC and a dimer of KaiA C-terminal domains from Thermosynechococcus elongatus | | Descriptor: | Circadian clock protein KaiA, Circadian clock protein KaiB, Circadian clock protein kinase KaiC, ... | | Authors: | Tseng, R, Goularte, N.F, Chavan, A, Luu, J, Chang, Y.G, Heilser, J, Tripathi, S, LiWang, A, Partch, C.L. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of the day-night transition in a bacterial circadian clock.
Science, 355, 2017
|
|
1A9E
 
 | | DECAMER-LIKE CONFORMATION OF A NANO-PEPTIDE BOUND TO HLA-B3501 DUE TO NONSTANDARD POSITIONING OF THE C-TERMINUS | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, B-35 B*3501 (ALPHA CHAIN), ... | | Authors: | Menssen, R, Orth, P, Ziegler, A, Saenger, W. | | Deposit date: | 1998-04-05 | | Release date: | 1998-10-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Decamer-like conformation of a nona-peptide bound to HLA-B*3501 due to non-standard positioning of the C terminus.
J.Mol.Biol., 285, 1999
|
|
1A9B
 
 | | DECAMER-LIKE CONFORMATION OF A NANO-PEPTIDE BOUND TO HLA-B3501 DUE TO NONSTANDARD POSITIONING OF THE C-TERMINUS | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, B-35 B*3501 (ALPHA CHAIN), ... | | Authors: | Menssen, R, Orth, P, Ziegler, A, Saenger, W. | | Deposit date: | 1998-04-03 | | Release date: | 1998-10-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Decamer-like conformation of a nona-peptide bound to HLA-B*3501 due to non-standard positioning of the C terminus.
J.Mol.Biol., 285, 1999
|
|
2ZIT
 
 | | Structure of the eEF2-ExoA-NAD+ complex | | Descriptor: | Elongation factor 2, Exotoxin A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jorgensen, R, Merrill, A.R. | | Deposit date: | 2008-02-24 | | Release date: | 2008-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The nature and character of the transition state for the ADP-ribosyltransferase reaction.
Embo Rep., 9, 2008
|
|
3B82
 
 | | Structure of the eEF2-ExoA(E546H)-NAD+ complex | | Descriptor: | Elongation factor 2, Exotoxin A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jorgensen, R, Merrill, A.R. | | Deposit date: | 2007-10-31 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The nature and character of the transition state for the ADP-ribosyltransferase reaction.
Embo Rep., 9, 2008
|
|
3B8H
 
 | | Structure of the eEF2-ExoA(E546A)-NAD+ complex | | Descriptor: | Elongation factor 2, Exotoxin A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jorgensen, R, Merrill, A.R. | | Deposit date: | 2007-11-01 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The nature and character of the transition state for the ADP-ribosyltransferase reaction.
Embo Rep., 9, 2008
|
|
3B78
 
 | | Structure of the eEF2-ExoA(R551H)-NAD+ complex | | Descriptor: | Elongation factor 2, Exotoxin A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jorgensen, R, Merrill, A.R. | | Deposit date: | 2007-10-30 | | Release date: | 2008-06-17 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The nature and character of the transition state for the ADP-ribosyltransferase reaction.
Embo Rep., 9, 2008
|
|
1U2R
 
 | | Crystal Structure of ADP-ribosylated Ribosomal Translocase from Saccharomyces cerevisiae | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jorgensen, R, Yates, S.P, Nilsson, J, Prentice, G.A, Teal, D.J, Merrill, A.R, Andersen, G.R. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of ADP-ribosylated Ribosomal Translocase from Saccharomyces cerevisiae
J.Biol.Chem., 279, 2004
|
|
