8B0W
 
 | |
8B0Z
 
 | |
7LTA
 
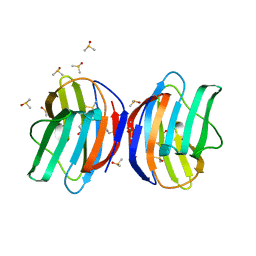 | | Galectin-1 in complex with Trehalose | | Descriptor: | DIMETHYL SULFOXIDE, Galectin-1, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Grimm, C, Bechold, J, Seibel, J. | | Deposit date: | 2021-02-19 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Galectin-1 in complex with Trehalose
To Be Published
|
|
7NML
 
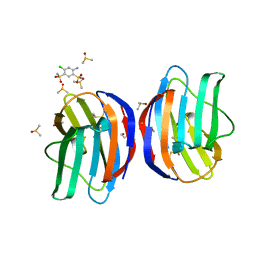 | | Galectin-1 in complex with 4-Amino-6-chloro-1,3-benzenedisulfonamide | | Descriptor: | 4-AMINO-6-CHLOROBENZENE-1,3-DISULFONAMIDE, DIMETHYL SULFOXIDE, Galectin-1 | | Authors: | Grimm, C, Bechold, J, Seibel, J. | | Deposit date: | 2021-02-23 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Galectin-1 in complex with 4-Amino-6-chloro-1,3-benzenedisulfonamide
To Be Published
|
|
3OM4
 
 | | Crystal structure of B. megaterium levansucrase mutant K373A | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Levansucrase, ... | | Authors: | Strube, C.P, Homann, A, Gamer, M, Jahn, D, Seibel, J, Heinz, D.W. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of B. megaterium levansucrase mutant K373A
To be Published
|
|
3OM2
 
 | | Crystal structure of B. megaterium levansucrase mutant D257A | | Descriptor: | CALCIUM ION, CITRIC ACID, Levansucrase, ... | | Authors: | Strube, C.P, Homann, A, Gamer, M, Jahn, D, Seibel, J, Heinz, D.W. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Polysaccharide Synthesis of the Levansucrase SacB from Bacillus megaterium Is Controlled by Distinct Surface Motifs.
J.Biol.Chem., 286, 2011
|
|
3OM5
 
 | | Crystal structure of B. megaterium levansucrase mutant N252A | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Strube, C.P, Homann, A, Gamer, M, Jahn, D, Seibel, J, Heinz, D.W. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Polysaccharide Synthesis of the Levansucrase SacB from Bacillus megaterium Is Controlled by Distinct Surface Motifs.
J.Biol.Chem., 286, 2011
|
|
3OM6
 
 | | Crystal structure of B. megaterium levansucrase mutant Y247A | | Descriptor: | CALCIUM ION, Levansucrase, SULFATE ION, ... | | Authors: | Strube, C.P, Homann, A, Gamer, M, Jahn, D, Seibel, J, Heinz, D.W. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Polysaccharide Synthesis of the Levansucrase SacB from Bacillus megaterium Is Controlled by Distinct Surface Motifs.
J.Biol.Chem., 286, 2011
|
|
3OM7
 
 | | Crystal structure of B. megaterium levansucrase mutant Y247W | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Levansucrase, ... | | Authors: | Strube, C.P, Homann, A, Gamer, M, Jahn, D, Seibel, J, Heinz, D.W. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Polysaccharide Synthesis of the Levansucrase SacB from Bacillus megaterium Is Controlled by Distinct Surface Motifs.
J.Biol.Chem., 286, 2011
|
|
6PWQ
 
 | | Crystal structure of Levansucrase from Bacillus subtilis mutant S164A at 2.6 A | | Descriptor: | CALCIUM ION, GLYCEROL, Glycoside hydrolase family 68 protein, ... | | Authors: | Diaz-Vilchis, A, Rodriguez-Alegria, M.E, Ortiz-Soto, M.E, Rudino-Pinera, E, Lopez-Munguia, A. | | Deposit date: | 2019-07-23 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Implications of the mutation S164A on Bacillus subtilis levansucrase product specificity and insights into protein interactions acting upon levan synthesis.
Int.J.Biol.Macromol., 161, 2020
|
|
5C8B
 
 | |
5MAN
 
 | |
5M9X
 
 | | Structure of sucrose phosphorylase from Bifidobacterium adolescentis bound to glycosylated resveratrol | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-[3-[(~{E})-2-(4-hydroxyphenyl)ethenyl]-5-oxidanyl-phenoxy]oxane-3,4,5-triol, Sucrose phosphorylase | | Authors: | Grimm, C, Kraus, M. | | Deposit date: | 2016-11-02 | | Release date: | 2017-12-20 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Switching enzyme specificity from phosphate to resveratrol glucosylation.
Chem. Commun. (Camb.), 53, 2017
|
|
2WA3
 
 | | FACTOR INHIBITING HIF-1 ALPHA WITH 2-(3-hydroxyphenyl)-2-oxoacetic acid | | Descriptor: | 2-(3-HYDROXYPHENYL)-2-OXO-ETHANOIC ACID, FE (II) ION, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR | | Authors: | Conejo-Garcia, A, Lienard, B.M.R, Clifton, I.J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2009-02-02 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for binding of cyclic 2-oxoglutarate analogues to factor-inhibiting hypoxia-inducible factor.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
2WA4
 
 | | FACTOR INHIBITING HIF-1 ALPHA WITH N,3-dihydroxybenzamide | | Descriptor: | FE (II) ION, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, N,3-DIHYDROXYBENZAMIDE, ... | | Authors: | Conejo-Garcia, A, Lienard, B.M.R, Clifton, I.J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2009-02-02 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for binding of cyclic 2-oxoglutarate analogues to factor-inhibiting hypoxia-inducible factor.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
