3ZK2
 
 | |
3ZK1
 
 | | Crystal structure of the sodium binding rotor ring at pH 5.3 | | Descriptor: | ATP SYNTHASE SUBUNIT C, DECYL-BETA-D-MALTOPYRANOSIDE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Schulz, S, Meier, T, Yildiz, O. | | Deposit date: | 2013-01-21 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A New Type of Na(+)-Driven ATP Synthase Membrane Rotor with a Two-Carboxylate Ion-Coupling Motif.
Plos Biol., 11, 2013
|
|
2WSX
 
 | | Crystal Structure of Carnitine Transporter from Escherichia coli | | Descriptor: | 3-CARBOXY-N,N,N-TRIMETHYLPROPAN-1-AMINIUM, L-CARNITINE/GAMMA-BUTYROBETAINE ANTIPORTER | | Authors: | Schulze, S, Terwisscha van Scheltinga, A.C, Kuehlbrandt, W. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Na(+)-Independent and Cooperative Substrate/Product Antiport in Cait.
Nature, 467, 2010
|
|
2WSW
 
 | | Crystal Structure of Carnitine Transporter from Proteus mirabilis | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, BCCT FAMILY BETAINE/CARNITINE/CHOLINE TRANSPORTER, GLYCEROL, ... | | Authors: | Schulze, S, Terwisscha van Scheltinga, A.C, Kuehlbrandt, W. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Structural Basis of Na(+)-Independent and Cooperative Substrate/Product Antiport in Cait.
Nature, 467, 2010
|
|
6HTY
 
 | | PXR in complex with P2X4 inhibitor compound 25 | | Descriptor: | (2~{R})-~{N}-[4-(3-chloranylphenoxy)-3-sulfamoyl-phenyl]-2-phenyl-propanamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hillig, R.C, Puetter, V, Werner, S, Mesch, S, Laux-Biehlmann, A, Braeuer, N, Dahloef, H, Klint, J, ter Laak, A, Pook, E, Neagoe, I, Nubbemeyer, R, Schulz, S. | | Deposit date: | 2018-10-05 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery and Characterization of the Potent and Selective P2X4 InhibitorN-[4-(3-Chlorophenoxy)-3-sulfamoylphenyl]-2-phenylacetamide (BAY-1797) and Structure-Guided Amelioration of Its CYP3A4 Induction Profile.
J.Med.Chem., 62, 2019
|
|
1WA9
 
 | | Crystal Structure of the PAS repeat region of the Drosophila clock protein PERIOD | | Descriptor: | PERIOD CIRCADIAN PROTEIN | | Authors: | Yildiz, O, Doi, M, Yujnovsky, I, Cardone, L, Berndt, A, Hennig, S, Schulze, S, Urbanke, C, Sassone-Corsi, P, Wolf, E. | | Deposit date: | 2004-10-25 | | Release date: | 2005-01-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structure and Interactions of the Pas Repeat Region of the Drosophila Clock Protein Period
Mol.Cell, 17, 2005
|
|
6Z1B
 
 | |
5G3U
 
 | | The structure of the L-tryptophan oxidase VioA from Chromobacterium violaceum in complex with its inhibitor 2-(1H-indol-3-ylmethyl)prop-2- enoic acid | | Descriptor: | 2-[(1H-indol-3-yl)methyl]prop-2-enoic acid, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Krausze, J, Rabe, J, Moser, J. | | Deposit date: | 2016-05-01 | | Release date: | 2016-08-03 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.377 Å) | | Cite: | Biosynthesis of Violacein, Structure and Function of l-Tryptophan Oxidase VioA from Chromobacterium violaceum.
J.Biol.Chem., 291, 2016
|
|
6TFI
 
 | | PXR IN COMPLEX WITH THROMBIN INHIBITOR COMPOUND 17 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hillig, R.C, Puetter, V. | | Deposit date: | 2019-11-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
4M8J
 
 | | Crystal structure of CaiT R262E bound to gamma-butyrobetaine | | Descriptor: | 3-CARBOXY-N,N,N-TRIMETHYLPROPAN-1-AMINIUM, L-carnitine/gamma-butyrobetaine antiporter | | Authors: | Kalayil, S. | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.294 Å) | | Cite: | Arginine oscillation explains Na+ independence in the substrate/product antiporter CaiT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6ZUG
 
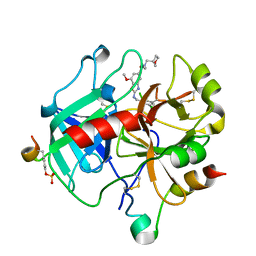 | | Crystal Structure of Thrombin in complex with compound10 | | Descriptor: | 2-[(3-chlorophenyl)methylamino]-7-methoxy-~{N}-[[(3~{S})-oxolan-3-yl]methyl]-~{N}-propyl-1,3-benzoxazole-5-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin-2, ... | | Authors: | Schafer, M. | | Deposit date: | 2020-07-22 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
6ZUU
 
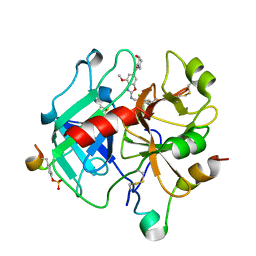 | | Crystal structure of Thrombin in complex with compound30 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin-2, Prothrombin, ... | | Authors: | Schafer, M. | | Deposit date: | 2020-07-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
6ZV7
 
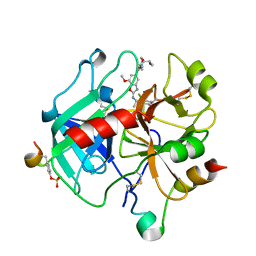 | | Crystal Structure of Thrombin in complex with compound42b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin-2, Prothrombin, ... | | Authors: | Schafer, M. | | Deposit date: | 2020-07-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
6ZV8
 
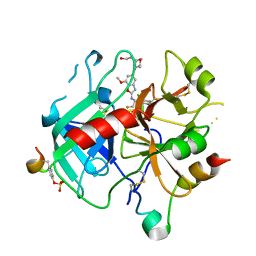 | | Crystal Structure of Thrombin in complex with compound51 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin-2, Prothrombin, ... | | Authors: | Schafer, M. | | Deposit date: | 2020-07-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
6ZUN
 
 | | Crystal Structure of Thrombin in complex with compound20a | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin-2, Prothrombin, ... | | Authors: | Schafer, M. | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
6ZUX
 
 | | Crystal Structure of Thrombin in complex with compound42a | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin-2, Prothrombin, ... | | Authors: | Schafer, M. | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
6ZUH
 
 | | Crystal Structure of Thrombin in complex with compound17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Hirudin-2, ... | | Authors: | Schafer, M. | | Deposit date: | 2020-07-22 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
6ZUW
 
 | | Crystal Structure of Thrombin in complex with compound40 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin-2, Prothrombin, ... | | Authors: | Schafer, M. | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
5G3T
 
 | | The structure of the L-tryptophan oxidase VioA from Chromobacterium violaceum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Krausze, J, Rabe, J, Moser, J. | | Deposit date: | 2016-05-01 | | Release date: | 2016-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biosynthesis of Violacein: Structure and Function of L-Tryptophan Oxidase Vioa Chromobacterium Violaceum
J.Biol.Chem., 291, 2016
|
|
5G3S
 
 | | The structure of the L-tryptophan oxidase VioA from Chromobacterium violaceum - Samarium derivative | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Krausze, J, Rabe, J, Moser, J. | | Deposit date: | 2016-05-01 | | Release date: | 2016-08-03 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (2.076 Å) | | Cite: | Biosynthesis of Violacein: Structure and Function of L-Tryptophan Oxidase Vioa Chromobacterium Violaceum
J.Biol.Chem., 291, 2016
|
|
8V2E
 
 | |
8SX3
 
 | | 10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10 | | Descriptor: | 10E8 Fab heavy chain, 10E8 light chain, 10E8-GT10.2 immunogen, ... | | Authors: | Huang, J, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-05-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Vaccination induces broadly neutralizing antibody precursors to HIV gp41.
Nat.Immunol., 25, 2024
|
|
8U03
 
 | |
8TZN
 
 | |
8TZW
 
 | |
