6BRE
 
 | | Hen Egg-White Lysozyme cocrystallized with Cadmium sulphate using CuKalpha source. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Sakon, J, Caviness, P, Jack, T, Pickens, J.B, Reed, P, Ruth, C. | | Deposit date: | 2017-11-30 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Hen Egg-White Lysozyme (HEWL) cocrystallized with Cadmium sulphate.
To Be Published
|
|
3TF4
 
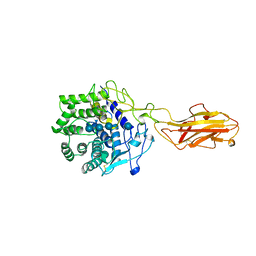 | | ENDO/EXOCELLULASE:CELLOTRIOSE FROM THERMOMONOSPORA | | Descriptor: | CALCIUM ION, T. FUSCA ENDO/EXO-CELLULASE E4 CATALYTIC DOMAIN AND CELLULOSE-BINDING DOMAIN, beta-D-glucopyranose, ... | | Authors: | Sakon, J, Wilson, D.B, Karplus, P.A. | | Deposit date: | 1997-05-30 | | Release date: | 1997-09-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca.
Nat.Struct.Biol., 4, 1997
|
|
1TF4
 
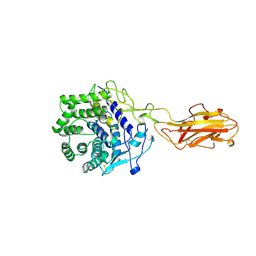 | | ENDO/EXOCELLULASE FROM THERMOMONOSPORA | | Descriptor: | CALCIUM ION, T. FUSCA ENDO/EXO-CELLULASE E4 CATALYTIC DOMAIN AND CELLULOSE-BINDING DOMAIN | | Authors: | Sakon, J, Wilson, D.B, Karplus, P.A. | | Deposit date: | 1997-05-30 | | Release date: | 1997-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca.
Nat.Struct.Biol., 4, 1997
|
|
4TF4
 
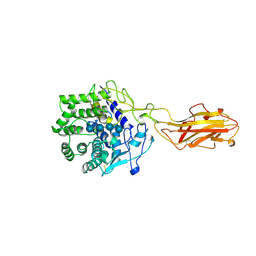 | | ENDO/EXOCELLULASE:CELLOPENTAOSE FROM THERMOMONOSPORA | | Descriptor: | CALCIUM ION, T. FUSCA ENDO/EXO-CELLULASE E4 CATALYTIC DOMAIN AND CELLULOSE-BINDING DOMAIN, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Sakon, J, Wilson, D.B, Karplus, P.A. | | Deposit date: | 1997-05-31 | | Release date: | 1997-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca.
Nat.Struct.Biol., 4, 1997
|
|
1JS4
 
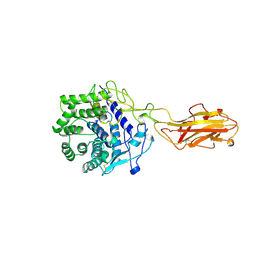 | | ENDO/EXOCELLULASE:CELLOBIOSE FROM THERMOMONOSPORA | | Descriptor: | CALCIUM ION, ENDO/EXOCELLULASE E4, beta-D-glucopyranose, ... | | Authors: | Sakon, J, Wilson, D.B, Karplus, P.A. | | Deposit date: | 1997-05-30 | | Release date: | 1997-09-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca.
Nat.Struct.Biol., 4, 1997
|
|
1ECE
 
 | | ACIDOTHERMUS CELLULOLYTICUS ENDOCELLULASE E1 CATALYTIC DOMAIN IN COMPLEX WITH A CELLOTETRAOSE | | Descriptor: | ENDOCELLULASE E1, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Sakon, J, Thomas, S.R, Himmel, M.E, Karplus, P.A. | | Deposit date: | 1996-04-04 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of thermostable family 5 endocellulase E1 from Acidothermus cellulolyticus in complex with cellotetraose.
Biochemistry, 35, 1996
|
|
1KAN
 
 | |
3JQW
 
 | | Crystal structure of Clostridium histolyticum colH collagenase collagen-binding domain 3 at 2 Angstrom resolution in presence of calcium | | Descriptor: | CALCIUM ION, ColH protein | | Authors: | Sakon, J, Philominathan, S.T.L, Matsushita, O, Bauer, R. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Comparison of ColH and ColG Collagen-Binding Domains from Clostridium histolyticum.
J.Bacteriol., 195, 2013
|
|
3JQX
 
 | | Crystal structure of Clostridium histolyticum colH collagenase collagen binding domain 3 at 2.2 Angstrom resolution in the presence of calcium and cadmium | | Descriptor: | CADMIUM ION, CALCIUM ION, ColH protein | | Authors: | Sakon, J, Philominathan, S.T.L, Bauer, R, Matsushita, O. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Comparison of ColH and ColG Collagen-Binding Domains from Clostridium histolyticum.
J.Bacteriol., 195, 2013
|
|
3EUU
 
 | |
4JRW
 
 | | Crystal structure of Clostridium histolyticum colg collagenase PKD domain 2 at 1.6 Angstrom resolution | | Descriptor: | BROMIDE ION, Collagenase | | Authors: | Sakon, J, Philominathan, S.T.L, Gann, S, Bauer, R, Matsushita, O. | | Deposit date: | 2013-03-22 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of three polycystic kidney disease-like domains from Clostridium histolyticum collagenases ColG and ColH.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5IKU
 
 | | Crystal structure of the Hathewaya histolytica ColG tandem collagen-binding domain s3as3b in the presence of calcium at 1.9 Angstrom resolution | | Descriptor: | CALCIUM ION, Collagenase | | Authors: | Janowska, K, Bauer, R, Roeser, R, Sakon, J, Matsushita, O. | | Deposit date: | 2016-03-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ca2+-induced orientation of tandem collagen binding domains from clostridial collagenase ColG permits two opposing functions of collagen fibril formation and retardation.
Febs J., 285, 2018
|
|
4U6T
 
 | | Crystal structure of the Clostridium histolyticum colH collagenase polycystic kidney disease-like domain 2a at 1.76 Angstrom resolution | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ColH protein | | Authors: | Bauer, R, Janowska, K, Sakon, J, Matsushita, O, Latimer, E. | | Deposit date: | 2014-07-29 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of three polycystic kidney disease-like domains from Clostridium histolyticum collagenases ColG and ColH.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4U7K
 
 | | Crystal structure of the Clostridium histolyticum colH collagenase polycystic kidney disease-like domain 2a in the presence of calcium at 1.9 Angstrom resolution | | Descriptor: | CALCIUM ION, ColH protein | | Authors: | Bauer, R, Janowska, K, Sakon, J, Matsushita, O, Latimer, E. | | Deposit date: | 2014-07-31 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structures of three polycystic kidney disease-like domains from Clostridium histolyticum collagenases ColG and ColH.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1VRX
 
 | | Endocellulase e1 from acidothermus cellulolyticus mutant y245g | | Descriptor: | ENDOCELLULASE E1 FROM A. CELLULOLYTICUS | | Authors: | Baker, J.O, McCarley, J.R, Lovett, R, Yu, C.H, Adney, W.S, Rignall, T.R, Vinzant, T.B, Decker, S.R, Sakon, J, Himmel, M.E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytically enhanced endocellulase Cel5A from Acidothermus cellulolyticus.
Appl.Biochem.Biotechnol., 121-124, 2005
|
|
2O8O
 
 | |
3CAF
 
 | |
3CU1
 
 | | Crystal Structure of 2:2:2 FGFR2D2:FGF1:SOS complex | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Fibroblast growth factor receptor 2, Heparin-binding growth factor 1 | | Authors: | Guo, F, Dakshinamurthy, R, Thallapuranam, S.K.K, Sakon, J. | | Deposit date: | 2008-04-15 | | Release date: | 2009-04-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of 2:2:2 FGFR2D2:FGF1:SOS complex
To be Published
|
|
1NQD
 
 | | CRYSTAL STRUCTURE OF CLOSTRIDIUM HISTOLYTICUM COLG COLLAGENASE COLLAGEN-BINDING DOMAIN 3B AT 1.65 ANGSTROM RESOLUTION IN PRESENCE OF CALCIUM | | Descriptor: | CALCIUM ION, class 1 collagenase | | Authors: | Wilson, J.J, Matsushita, O, Okabe, A, Sakon, J. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Bacterial Collagen-Binding Domain with Novel Calcium-Binding Motif Controls Domain Orientation
Embo J., 22, 2003
|
|
1NQJ
 
 | | CRYSTAL STRUCTURE OF CLOSTRIDIUM HISTOLYTICUM COLG COLLAGENASE COLLAGEN-BINDING DOMAIN 3B AT 1.0 ANGSTROM RESOLUTION IN ABSENCE OF CALCIUM | | Descriptor: | CHLORIDE ION, LITHIUM ION, class 1 collagenase | | Authors: | Wilson, J.J, Matsushita, O, Okabe, A, Sakon, J. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A bacterial collagen-binding domain with novel calcium-binding motif
controls domain orientation
Embo J., 22, 2003
|
|
4H7B
 
 | |
1II3
 
 | |
1R77
 
 | | Crystal structure of the cell wall targeting domain of peptidylglycan hydrolase ALE-1 | | Descriptor: | Cell Wall Targeting Domain of Glycylglycine Endopeptidase ALE-1 | | Authors: | Lu, J.Z, Fujiwara, T, Komatsuzawa, H, Sugai, M, Sakon, J. | | Deposit date: | 2003-10-20 | | Release date: | 2005-04-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cell Wall-targeting Domain of Glycylglycine Endopeptidase Distinguishes among Peptidoglycan Cross-bridges.
J.Biol.Chem., 281, 2006
|
|
1IHZ
 
 | |
4TN9
 
 | | Crystal structure of Clostridium histolyticum ColG collagenase polycystic kidney disease-like domain at 1.4 Angstrom resolution | | Descriptor: | Collagenase | | Authors: | Bauer, R, Sakon, J, Philominathan, S.T.L, Matsushita, O, Gann, S. | | Deposit date: | 2014-06-03 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of three polycystic kidney disease-like domains from Clostridium histolyticum collagenases ColG and ColH.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
