4CVB
 
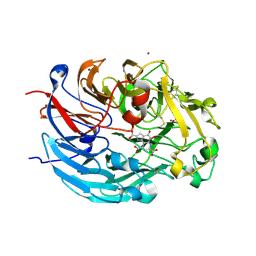 | | Crystal structure of quinone-dependent alcohol dehydrogenase from Pseudogluconobacter saccharoketogenenes | | Descriptor: | ALCOHOL DEHYDROGENASE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, Yu, S, Mikkelsen, R, Nikolaev, I, Mulder, H, Dijkstra, B.W. | | Deposit date: | 2014-03-25 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structure of Quinone-Dependent Alcohol Dehydrogenase from Pseudogluconobacter Saccharoketogenes. A Versatile Dehydrogenase Oxidizing Alcohols and Carbohydrates.
Protein Sci., 24, 2015
|
|
4CVC
 
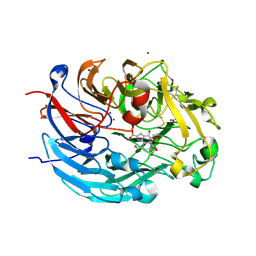 | | Crystal structure of quinone-dependent alcohol dehydrogenase from Pseudogluconobacter saccharoketogenenes with zinc in the active site | | Descriptor: | ALCOHOL DEHYDROGENASE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rozeboom, H.J, Yu, S, Mikkelsen, R, Nikolaev, I, Mulder, H, Dijkstra, B.W. | | Deposit date: | 2014-03-25 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Quinone-Dependent Alcohol Dehydrogenase from Pseudogluconobacter Saccharoketogenes. A Versatile Dehydrogenase Oxidizing Alcohols and Carbohydrates.
Protein Sci., 24, 2015
|
|
2X2J
 
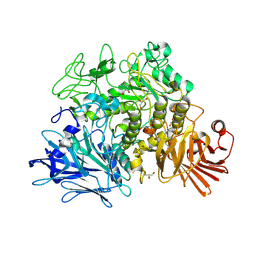 | | Crystal structure of the Gracilariopsis lemaneiformis alpha- 1,4-glucan lyase with deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
2PYG
 
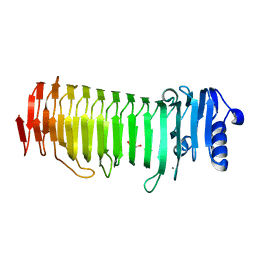 | | Azotobacter vinelandii Mannuronan C-5 epimerase AlgE4 A-module | | Descriptor: | CALCIUM ION, GLYCEROL, Poly(beta-D-mannuronate) C5 epimerase 4 | | Authors: | Rozeboom, H.J, Bjerkan, T.M, Kalk, K.H, Ertesvag, H, Holtan, S, Aachmann, F.L, Valla, S, Dijkstra, B.W. | | Deposit date: | 2007-05-16 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mutational Characterization of the Catalytic A-module of the Mannuronan C-5-epimerase AlgE4 from Azotobacter vinelandii.
J.Biol.Chem., 283, 2008
|
|
2PYH
 
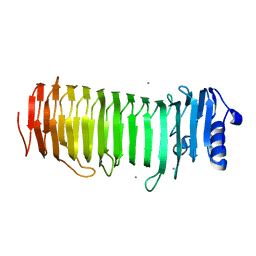 | | Azotobacter vinelandii Mannuronan C-5 epimerase AlgE4 A-module complexed with mannuronan trisaccharide | | Descriptor: | CALCIUM ION, Poly(beta-D-mannuronate) C5 epimerase 4, alpha-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid | | Authors: | Rozeboom, H.J, Bjerkan, T.M, Kalk, K.H, Ertesvag, H, Holtan, S, Aachman, F.L, Valla, S, Dijkstra, B.W. | | Deposit date: | 2007-05-16 | | Release date: | 2008-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mutational Characterization of the Catalytic A-module of the Mannuronan C-5-epimerase AlgE4 from Azotobacter vinelandii.
J.Biol.Chem., 283, 2008
|
|
2X2I
 
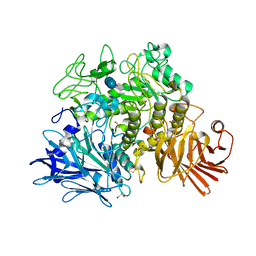 | | Crystal structure of the Gracilariopsis lemaneiformis alpha-1,4- glucan lyase with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, GLYCEROL | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
2X2H
 
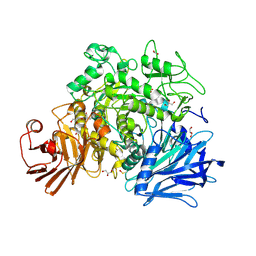 | | Crystal structure of the Gracilariopsis lemaneiformis alpha-1,4- glucan lyase | | Descriptor: | ACETATE ION, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
4AMX
 
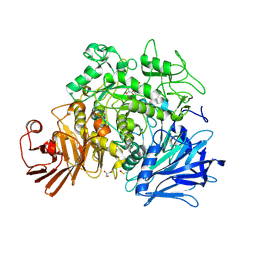 | | CRYSTAL STRUCTURE OF THE GRACILARIOPSIS LEMANEIFORMIS ALPHA-1,4- GLUCAN LYASE Covalent Intermediate Complex with 5-fluoro-glucosyl- fluoride | | Descriptor: | 5-fluoro-beta-D-glucopyranose, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, GLYCEROL, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2012-03-14 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
4AMW
 
 | | CRYSTAL STRUCTURE OF THE GRACILARIOPSIS LEMANEIFORMIS ALPHA-1,4- GLUCAN LYASE Covalent Intermediate Complex with 5-fluoro-idosyl- fluoride | | Descriptor: | 5-fluoro-alpha-L-idopyranose, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, GLYCEROL, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2012-03-14 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
4C2L
 
 | | Crystal structure of endo-xylogalacturonan hydrolase from Aspergillus tubingensis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-XYLOGALACTURONAN HYDROLASE A, ... | | Authors: | Rozeboom, H.J, Beldman, G, Schols, H.A, Dijkstra, B.W. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Endo-Xylogalacturonan Hydrolase from Aspergillus Tubingensis.
FEBS J., 280, 2013
|
|
4CCW
 
 | | Crystal structure of naproxen esterase (carboxylesterase NP) from Bacillus subtilis | | Descriptor: | (2-hydroxyethoxy)acetic acid, CARBOXYL ESTERASE NP | | Authors: | Rozeboom, H.J, Godinho, L.F, Nardini, M, Quax, W.J, Dijkstra, B.W. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Two Bacillus Carboxylesterases with Different Enantioselectivities.
Biochim.Biophys.Acta, 1844, 2014
|
|
4CCY
 
 | | Crystal structure of carboxylesterase CesB (YbfK) from Bacillus subtilis | | Descriptor: | CARBOXYLESTERASE YBFK, SODIUM ION | | Authors: | Rozeboom, H.J, Godinho, L.F, Nardini, M, Quax, W.J, Dijkstra, B.W. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structures of Two Bacillus Carboxylesterases with Different Enantioselectivities.
Biochim.Biophys.Acta, 1844, 2014
|
|
8QYG
 
 | | Crystal structure of Nitroreductase from Bacillus tequilensis | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Rozeboom, H.J, Russo, S, Fraaije, M.W, Poelarends, G.J. | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-08 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Biochemical, kinetic, and structural characterization of a Bacillus tequilensis nitroreductase.
Febs J., 2024
|
|
3BMW
 
 | | Cyclodextrin glycosyl transferase from Thermoanerobacterium thermosulfurigenes EM1 mutant S77P complexed with a maltoheptaose inhibitor | | Descriptor: | 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, van Oosterwijk, N, Dijkstra, B.W. | | Deposit date: | 2007-12-13 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elimination of competing hydrolysis and coupling side reactions of a cyclodextrin glucanotransferase by directed evolution.
Biochem.J., 413, 2008
|
|
8S1J
 
 | |
6I9G
 
 | |
6YRA
 
 | |
6Z1X
 
 | |
6Z1W
 
 | |
7AM3
 
 | | Crystal structure of Peptiligase mutant - M222P | | Descriptor: | GLYCEROL, SULFATE ION, Subtilisin BPN' | | Authors: | Rozeboom, H.J, Janssen, D.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | From thiol-subtilisin to omniligase: Design and structure of a broadly applicable peptide ligase.
Comput Struct Biotechnol J, 19, 2021
|
|
7AM6
 
 | |
7AM8
 
 | | Crystal structure of Omniligase mutant W189F | | Descriptor: | ACRYLIC ACID, CHLORIDE ION, HISTIDINE, ... | | Authors: | Rozeboom, H.J, Janssen, D.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | From thiol-subtilisin to omniligase: Design and structure of a broadly applicable peptide ligase.
Comput Struct Biotechnol J, 19, 2021
|
|
7AM4
 
 | |
7AM5
 
 | |
7AM7
 
 | |
