5MWU
 
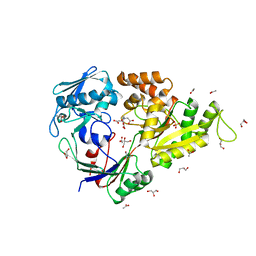 | | Crystal structure of the periplasmic nickel-binding protein NikA from Escherichia coli in complex with Ru(bpza)(CO)2Cl | | Descriptor: | ACETATE ION, CARBON MONOXIDE, CHLORIDE ION, ... | | Authors: | Cavazza, C, Lopez, S, Rondot, L, Iannello, M, Boeri-Erba, E, Burzlaff, N, Strinitz, F, Jorge-Robin, A, Marchi-Delapierre, C, Menage, S. | | Deposit date: | 2017-01-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efficient conversion of alkenes to chlorohydrins by a Ru-based artificial enzyme
To Be Published
|
|
6F5V
 
 | | Crystal structure of the prephenate aminotransferase from Arabidopsis thaliana | | Descriptor: | Bifunctional aspartate aminotransferase and glutamate/aspartate-prephenate aminotransferase, CITRIC ACID, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cobessi, D, Robin, A, Giustini, C, Graindorge, M, Matringe, M. | | Deposit date: | 2017-12-03 | | Release date: | 2019-03-13 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tyrosine metabolism: identification of a key residue in the acquisition of prephenate aminotransferase activity by 1 beta aspartate aminotransferase.
Febs J., 286, 2019
|
|
3L76
 
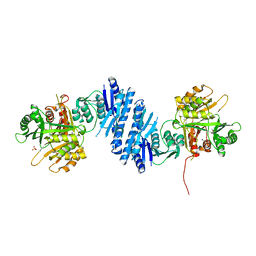 | | Crystal Structure of Aspartate Kinase from Synechocystis | | Descriptor: | Aspartokinase, LYSINE, SULFATE ION, ... | | Authors: | Robin, A, Cobessi, D, Curien, G, Robert-Genthon, M, Ferrer, J.-L, Dumas, R. | | Deposit date: | 2009-12-28 | | Release date: | 2010-06-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | A new mode of dimerization of allosteric enzymes with ACT domains revealed by the crystal structure of the aspartate kinase from Cyanobacteria
J.Mol.Biol., 399, 2010
|
|
7OEZ
 
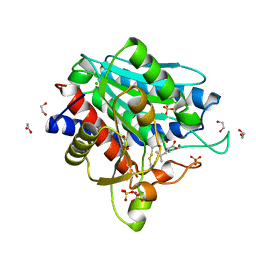 | | Leucine Aminopeptidase A mature enzyme in a complex with leucine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Watson, K.A, Baltulionis, G. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The role of propeptide-mediated autoinhibition and intermolecular chaperone in the maturation of cognate catalytic domain in leucine aminopeptidase.
J.Struct.Biol., 213, 2021
|
|
6ZEQ
 
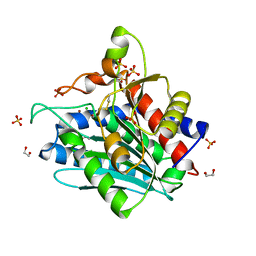 | | Aspergillus oryzae Leucine Aminopeptidase A mature enzyme | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Watson, K.A, Baltulionis, G. | | Deposit date: | 2020-06-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The role of propeptide-mediated autoinhibition and intermolecular chaperone in the maturation of cognate catalytic domain in leucine aminopeptidase.
J.Struct.Biol., 213, 2021
|
|
6ZEP
 
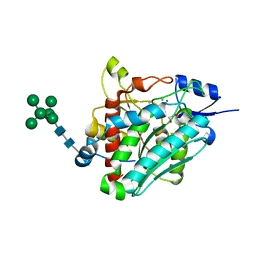 | | Flavourzyme Leucine Aminopeptidase A proenzyme | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Leucine aminopeptidase A, ... | | Authors: | Watson, K.A, Baltulionis, G. | | Deposit date: | 2020-06-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The role of propeptide-mediated autoinhibition and intermolecular chaperone in the maturation of cognate catalytic domain in leucine aminopeptidase.
J.Struct.Biol., 213, 2021
|
|
5L8D
 
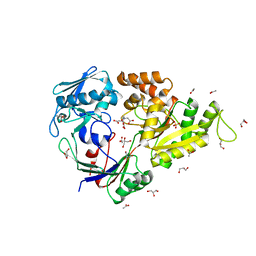 | |
4I9D
 
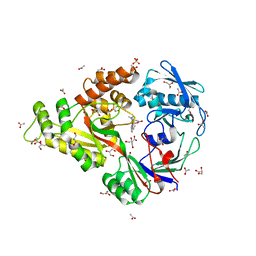 | | X-ray structure of NikA in complex with Fe-N,N'-Bis(2-pyridylmethyl)-N-carboxymethyl-N'-methyl | | Descriptor: | ACETATE ION, GLYCEROL, Nickel-binding periplasmic protein, ... | | Authors: | Cherrier, M.V, Amara, P, Iannello, M, Cavazza, C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An artificial oxygenase built from scratch: substrate binding site identified using a docking approach.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
7NAY
 
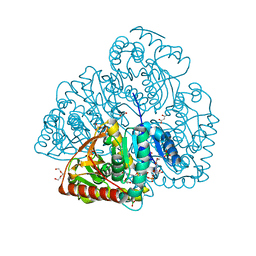 | | Crystal structure of lactate dehydrogenase from Selenomonas ruminantium with NADH. | | Descriptor: | 1,2-ETHANEDIOL, BORIC ACID, L-lactate dehydrogenase, ... | | Authors: | Bertrand, Q, Robin, A, Girard, E, Madern, D. | | Deposit date: | 2021-01-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Biochemical, structural and dynamical characterizations of the lactate dehydrogenase from Selenomonas ruminantium provide information about an intermediate evolutionary step prior to complete allosteric regulation acquisition in the super family of lactate and malate dehydrogenases.
J.Struct.Biol., 215, 2023
|
|
6F35
 
 | | Crystal structure of the aspartate aminotranferase from Rhizobium meliloti | | Descriptor: | ACETATE ION, Aspartate aminotransferase B, GLYCEROL, ... | | Authors: | Cobessi, D, Graindorge, M, Giustini, C, Matringe, M. | | Deposit date: | 2017-11-28 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tyrosine metabolism: identification of a key residue in the acquisition of prephenate aminotransferase activity by 1 beta aspartate aminotransferase.
Febs J., 286, 2019
|
|
6F77
 
 | | Crystal structure of the prephenate aminotransferase from Rhizobium meliloti | | Descriptor: | Aspartate aminotransferase A, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Cobessi, D, Giustini, C, Graindorge, M, Matringe, M. | | Deposit date: | 2017-12-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Tyrosine metabolism: identification of a key residue in the acquisition of prephenate aminotransferase activity by 1 beta aspartate aminotransferase.
Febs J., 286, 2019
|
|
