2I58
 
 | | Crystal Structure of RafE from Streptococcus pneumoniae complexed with raffinose | | Descriptor: | CHLORIDE ION, Sugar ABC transporter, sugar-binding protein, ... | | Authors: | Paterson, N.G, Riboldi-Tunnicliffe, A, Mitchell, T.J, Isaacs, N.W. | | Deposit date: | 2006-08-24 | | Release date: | 2007-08-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of apo and bound forms of RafE from Streptococcus pneumoniae
To be Published
|
|
3RPK
 
 | |
2HEU
 
 | | Atomic resolution structure of apo-form of RafE from Streptococcus pneumoniae | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Paterson, N.G, Riboldi-Tunnicliffe, A, Mitchell, T.J, Isaacs, N.W. | | Deposit date: | 2006-06-22 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | High resolution crystal structures of RafE from Streptococcus pneumoniae.
To be Published
|
|
2HFB
 
 | |
2HQ0
 
 | | Structure of RafE from Streptococcus pneumoniae | | Descriptor: | ACETATE ION, SODIUM ION, Sugar ABC transporter, ... | | Authors: | Paterson, N.G, Riboldi-Tunnicliffe, A, Mitchell, T.J, Isaacs, N.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High resolution crystal structures of RafE from Streptococcus pneumoniae
To be Published
|
|
4HSS
 
 | |
4HSQ
 
 | |
4TKO
 
 | | Structure of the periplasmic adaptor protein EmrA | | Descriptor: | EmrA, ISOPROPYL ALCOHOL, MAGNESIUM ION | | Authors: | Hinchliffe, P, Greene, N.P, Paterson, N.G, Crow, A, Hughes, C, Koronakis, V. | | Deposit date: | 2014-05-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of the periplasmic adaptor protein from a major facilitator superfamily (MFS) multidrug efflux pump.
Febs Lett., 588, 2014
|
|
8BFD
 
 | | Racemic structure of PK-7 (310HD-U2U5) | | Descriptor: | 310HD-U2U5, D-310HD-U2U5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
8BFE
 
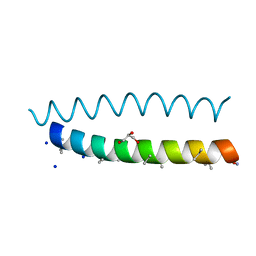 | | A dimeric de novo coiled-coil assembly: PK-2 (CC-TypeN-LaUbUcLd) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CC-TypeN-LaUbUcLd, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
4N4R
 
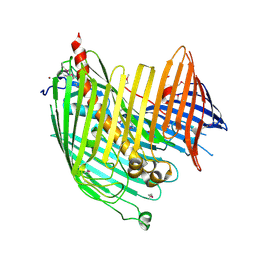 | | Structure basis of lipopolysaccharide biogenesis | | Descriptor: | CACODYLATE ION, LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ... | | Authors: | Dong, H, Xiang, Q, Wang, Z, Paterson, N.G, He, C, Zhang, Y, Wang, W, Dong, C. | | Deposit date: | 2013-10-08 | | Release date: | 2014-06-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for outer membrane lipopolysaccharide insertion.
Nature, 511, 2014
|
|
5FJE
 
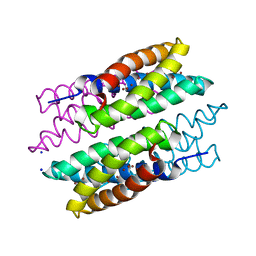 | | CU(I)-CSP1 (COPPER STORAGE PROTEIN 1) FROM METHYLOSINUS TRICHOSPORIUM OB3B | | Descriptor: | COPPER (I) ION, COPPER STORAGE PROTEIN 1, SODIUM ION | | Authors: | Vita, N, Platsaki, S, Basle, A, Allen, S.J, Paterson, N.G, Crombie, A.T, Murrell, J.C, Waldron, K.J, Dennison, C. | | Deposit date: | 2015-10-07 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Four-Helix Bundle Stores Copper for Methane Oxidation.
Nature, 525, 2015
|
|
5FJD
 
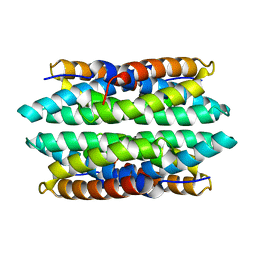 | | APO-CSP1 (COPPER STORAGE PROTEIN 1) FROM METHYLOSINUS TRICHOSPORIUM OB3B | | Descriptor: | COPPER STORAGE PROTEIN 1 | | Authors: | Vita, N, Platsaki, S, Basle, A, Allen, S.J, Paterson, N.G, Crombie, A.T, Murrell, J.C, Waldron, K.J, Dennison, C. | | Deposit date: | 2015-10-07 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Four-Helix Bundle Stores Copper for Methane Oxidation.
Nature, 525, 2015
|
|
8OYR
 
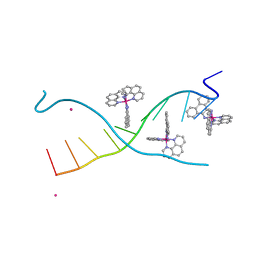 | | DNA Major Groove Binding by lambda-[Ru(phen)2(phi)]2+ | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3'), STRONTIUM ION, ruthenium polypyridyl complex (lambda enantiomer) | | Authors: | Prieto-Otoya, T.D, Cardin, C.J, McQuaid, K.T, Paterson, N.G. | | Deposit date: | 2023-05-05 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Probing a Major DNA Weakness: Resolving the Groove and Sequence Selectivity of the Diimine Complex Lambda-[Ru(phen) 2 phi] 2.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8QKS
 
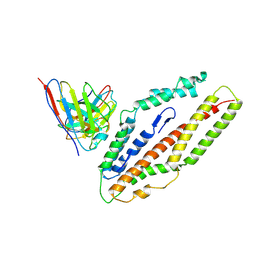 | | Plasmodium falciparum reticulocyte-binding protein homologue 5 (PfRH5) bound to R5.034 | | Descriptor: | Immunoglobulin lambda variable 1-36, R5034HV, Reticulocyte-binding protein-like protein 5 | | Authors: | Wright, N.D, Barrett, J.R, Bradshaw, W.J, Paterson, N.G, MacLean, E.M, Ferreira, L, McHugh, K, Von Delft, F, Koekemoer, L, Draper, S.J. | | Deposit date: | 2023-09-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.994 Å) | | Cite: | Analysis of the diverse antigenic landscape of the malaria protein RH5 identifies a potent vaccine-induced human public antibody clonotype.
Cell, 187, 2024
|
|
8QKR
 
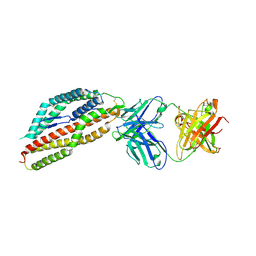 | | Plasmodium falciparum reticulocyte-binding protein homologue 5 (PfRH5) bound to R5.251 | | Descriptor: | R5251VHCH, R5251VLCL, Reticulocyte-binding protein-like protein 5 | | Authors: | Wright, N.D, Barrett, J.R, Bradshaw, W.J, Paterson, N.G, MacLean, E.M, Ferreira, L, McHugh, K, Koekemoer, L, Draper, S.J. | | Deposit date: | 2023-09-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.234 Å) | | Cite: | Analysis of the diverse antigenic landscape of the malaria protein RH5 identifies a potent vaccine-induced human public antibody clonotype.
Cell, 187, 2024
|
|
8RLY
 
 | | E. coli endonuclease IV complexed with sulfate, catalytic Fe2+ | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Endonuclease 4, ... | | Authors: | Saper, M.A, Paterson, N.G, Kirillov, S, Rouvinski, A. | | Deposit date: | 2024-01-04 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Octahedral Iron in Catalytic Sites of Endonuclease IV from Staphylococcus aureus and Escherichia coli .
Biochemistry, 64, 2025
|
|
2XE4
 
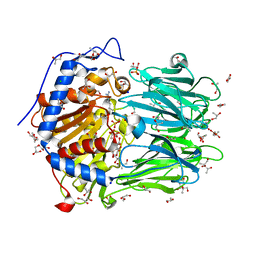 | | Structure of Oligopeptidase B from Leishmania major | | Descriptor: | ANTIPAIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | McLuskey, K, Paterson, N.G, Bland, N.D, Mottram, J.C, Isaacs, N.W. | | Deposit date: | 2010-05-11 | | Release date: | 2010-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Leishmania Major Oligopeptidase B Gives Insight Into the Enzymatic Properties of a Trypanosomatid Virulence Factor.
J.Biol.Chem., 285, 2010
|
|
7OTB
 
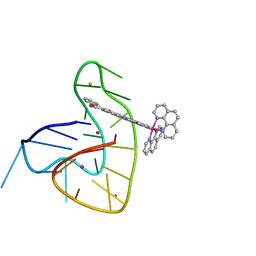 | | Ruthenium polypridyl complex bound to a unimolecular chair-form G-quadruplex | | Descriptor: | BARIUM ION, DNA (5'-D(*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*TP*GP*GP*G)-3'), POTASSIUM ION, ... | | Authors: | McQuaid, K.T, Cardin, C.J, Hall, J.P, Paterson, N.G, Baumgaertner, L. | | Deposit date: | 2021-06-09 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ruthenium Polypyridyl Complex Bound to a Unimolecular Chair-Form G-Quadruplex.
J.Am.Chem.Soc., 144, 2022
|
|
3HTL
 
 | | Structure of the Corynebacterium diphtheriae major pilin SpaA points to a modular pilus assembly with stabilizing isopeptide bonds | | Descriptor: | CALCIUM ION, Putative surface-anchored fimbrial subunit, SODIUM ION | | Authors: | Kang, H.J, Paterson, N.G, Gaspar, A.H, Ton-That, H, Baker, E.N. | | Deposit date: | 2009-06-11 | | Release date: | 2009-09-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HR6
 
 | | Structure of the Corynebacterium diphtheriae major pilin SpaA points to a modular pilus assembly stabilizing isopeptide bonds | | Descriptor: | CALCIUM ION, IODIDE ION, Putative surface-anchored fimbrial subunit | | Authors: | Kang, H.J, Paterson, N.G, Gaspar, A.H, Ton-That, H, Baker, E.N. | | Deposit date: | 2009-06-08 | | Release date: | 2009-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5LRN
 
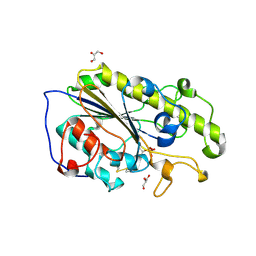 | | Structure of mono-zinc MCR-1 in P21 space group | | Descriptor: | GLYCEROL, Phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Hinchliffe, P, Paterson, N.G, Spencer, J. | | Deposit date: | 2016-08-19 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Insights into the Mechanistic Basis of Plasmid-Mediated Colistin Resistance from Crystal Structures of the Catalytic Domain of MCR-1.
Sci Rep, 7, 2017
|
|
7QDI
 
 | | Structure of octameric left-handed 310-helix bundle: D-310HD | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2021-11-27 | | Release date: | 2022-04-27 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
7QDK
 
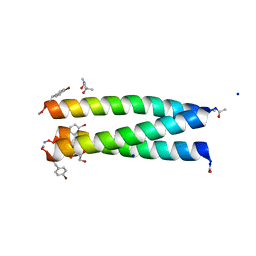 | | A trimeric de novo coiled-coil assembly: CC-TypeN-LaLd | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CC-TypeN-LaLd, GLYCEROL, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2021-11-27 | | Release date: | 2022-04-27 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
7QDJ
 
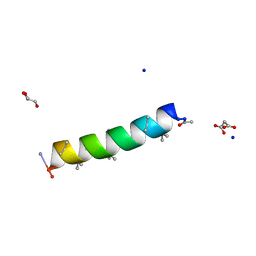 | | Racemic structure of PK-10 and PK-11 | | Descriptor: | GLYCEROL, MALONATE ION, PK-10+PK-11, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2021-11-27 | | Release date: | 2022-04-27 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
