2N37
 
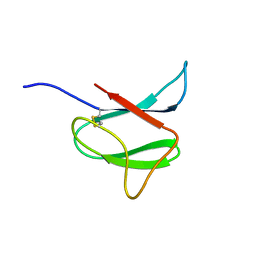 | | Solution structure of AVR-Pia | | Descriptor: | AVR-Pia protein | | Authors: | Ose, T, Oikawa, A, Nakamura, Y, Maenaka, K, Higuchi, Y, Satoh, Y, Fujiwara, S, Demura, M, Sone, T. | | Deposit date: | 2015-05-25 | | Release date: | 2015-10-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an avirulence protein, AVR-Pia, from Magnaporthe oryzae
J.Biomol.Nmr, 63, 2015
|
|
1J0D
 
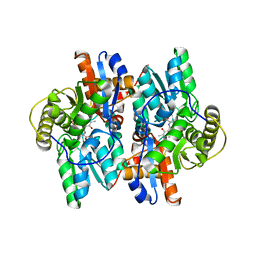 | | ACC deaminase mutant complexed with ACC | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
1J0E
 
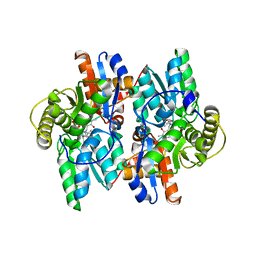 | | ACC deaminase mutant reacton intermediate | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
1IZC
 
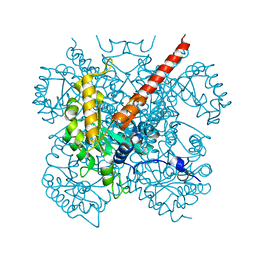 | | Crystal Structure Analysis of Macrophomate synthase | | Descriptor: | MAGNESIUM ION, PYRUVIC ACID, macrophomate synthase intermolecular Diels-Alderase | | Authors: | Ose, T, Watanabe, K, Mie, T, Honma, M, Watanabe, H, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2002-10-01 | | Release date: | 2003-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insight into a natural Diels-Alder reaction from the structure of macrophomate synthase.
Nature, 422, 2003
|
|
1V43
 
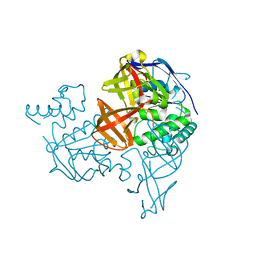 | | Crystal Structure of ATPase subunit of ABC Sugar Transporter | | Descriptor: | sugar-binding transport ATP-binding protein | | Authors: | Ose, T, Fujie, T, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-11-08 | | Release date: | 2004-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the ATP-binding cassette of multisugar transporter from Pyrococcus horikoshii OT3
Proteins, 57, 2004
|
|
1VCI
 
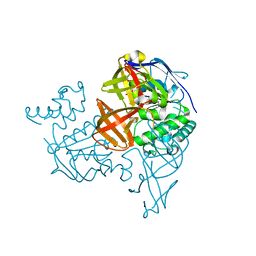 | | Crystal structure of the ATP-binding cassette of multisugar transporter from Pyrococcus horikoshii OT3 complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, sugar-binding transport ATP-binding protein | | Authors: | Ose, T, Fujie, T, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2004-03-08 | | Release date: | 2004-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the ATP-binding cassette of multisugar transporter from Pyrococcus horikoshii OT3
Proteins, 57, 2004
|
|
2UWM
 
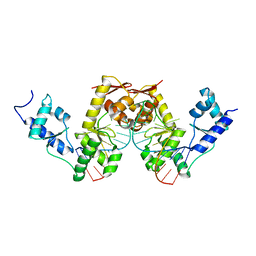 | | C-TERMINAL DOMAIN(WH2-WH4) OF ELONGATION FACTOR SELB IN COMPLEX WITH SECIS RNA | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*UP*GP*CP*CP*GP *GP*UP*CP*UP*GP*GP*CP*AP*AP*CP*GP*CP*C)-3', SELENOCYSTEINE-SPECIFIC ELONGATION FACTOR | | Authors: | Ose, T, Soler, N, Rasubala, L, Kuroki, K, Kohda, D, Fourmy, D, Yoshizawa, S, Maenaka, K. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis for Dynamic Interdomain Movement and RNA Recognition of the Selenocysteine-Specific Elongation Factor Selb.
Structure, 15, 2007
|
|
1J0C
 
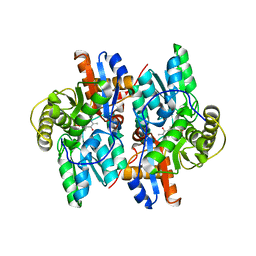 | | ACC deaminase mutated to catalytic residue | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
1WE5
 
 | | Crystal Structure of Alpha-Xylosidase from Escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative family 31 glucosidase yicI | | Authors: | Ose, T, Kitamura, M, Okuyama, M, Mori, H, Kimura, A, Watanabe, N, Yao, M, Tanaka, I. | | Deposit date: | 2004-05-24 | | Release date: | 2005-02-15 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Alpha-Xylosidase from Escherichia coli
TO BE PUBLISHED
|
|
3A3Q
 
 | | Structure of N59D HEN EGG-WHITE LYSOZYME in complex with (GlcNAc)3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme C | | Authors: | Ose, T, Kuroki, K, Matsushima, M, Maenaka, K, Kumagai, I. | | Deposit date: | 2009-06-16 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Importance of the hydrogen bonding network including Asp52 for catalysis, as revealed by Asn59 mutant hen egg-white lysozymes
J.Biochem., 146, 2009
|
|
3A3R
 
 | | Structure of N59D HEN EGG-WHITE LYSOZYME | | Descriptor: | Lysozyme C | | Authors: | Ose, T, Kuroki, K, Matsushima, M, Maenaka, K, Kumagai, I. | | Deposit date: | 2009-06-16 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Importance of the hydrogen bonding network including Asp52 for catalysis, as revealed by Asn59 mutant hen egg-white lysozymes
J.Biochem., 146, 2009
|
|
3X1O
 
 | | Crystal structure of the ROQ domain of human Roquin | | Descriptor: | IODIDE ION, Roquin-1 | | Authors: | Ose, T, Verma, A, Cockburn, J.B, Berrow, N.S, Alderton, D, Stuart, D, Owens, R.J, Jones, E.Y. | | Deposit date: | 2014-11-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Roquin binds microRNA-146a and Argonaute2 to regulate microRNA homeostasis
Nat Commun, 6, 2015
|
|
8IAY
 
 | | Crystal structure of canine distemper virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin glycoprotein | | Authors: | Fukuhara, H, Yumoto, K, Sako, M, Kajikawa, M, Ose, T, Hashiguchi, T, Kamishikiryo, J, Maita, N, Kuroki, K, Maenaka, K. | | Deposit date: | 2023-02-09 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.027 Å) | | Cite: | Glycan-shielded homodimer structure and dynamical features of the canine distemper virus hemagglutinin relevant for viral entry and efficient vaccination
Elife, 2024
|
|
4WCO
 
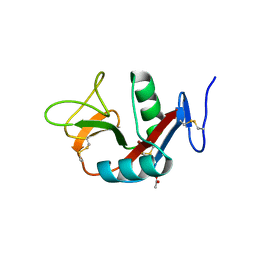 | | Crystal structure of extracellular domain of human lectin-like transcript 1 (LLT1), the ligand for natural killer receptor-P1A | | Descriptor: | ACETATE ION, C-type lectin domain family 2 member D, SULFATE ION, ... | | Authors: | Kita, S, Matsubara, H, Kasai, Y, Tamaoki, T, Okabe, Y, Fukuhara, H, Kamishikiryo, J, Ose, T, Kuroki, K, Maenaka, K. | | Deposit date: | 2014-09-05 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure of extracellular domain of human lectin-like transcript 1 (LLT1), the ligand for natural killer receptor-P1A
Eur.J.Immunol., 45, 2015
|
|
1F2D
 
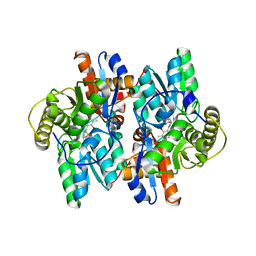 | | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE DEAMINASE | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE DEAMINASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Yao, M, Ose, T, Sugimoto, H, Horiuchi, A, Nakagawa, A, Yokoi, D, Murakami, T, Honma, M, Wakatsuki, S, Tanaka, I. | | Deposit date: | 2000-05-24 | | Release date: | 2000-12-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 1-aminocyclopropane-1-carboxylate deaminase from Hansenula saturnus.
J.Biol.Chem., 275, 2000
|
|
8HRH
 
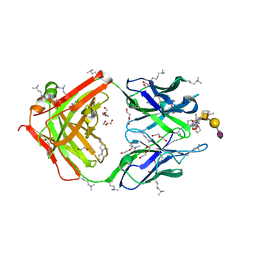 | | SN-131/1B2 anti-MUC1 antibody with a glycopeptide | | Descriptor: | 1-ACETYL-L-PROLINE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALANINE, ... | | Authors: | Wakui, H, Horidome, C, Yao, M, Ose, T, Nishimura, S.-I. | | Deposit date: | 2022-12-15 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural and molecular insight into antibody recognition of dynamic neoepitopes in membrane tethered MUC1 of pancreatic cancer cells and secreted exosomes.
Rsc Chem Biol, 4, 2023
|
|
2V1T
 
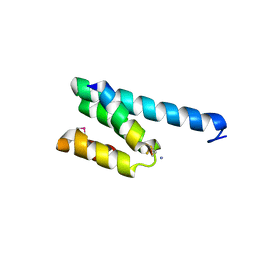 | | CRYSTAL STRUCTURE OF RAT TOM20-ALDH PRESEQUENCE COMPLEX | | Descriptor: | ALDEHYDE DEHYDROGENASE, MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20 HOMOLOG | | Authors: | Obita, T, Igura, M, Ose, T, Endo, T, Maenaka, K, Kohda, D. | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Tom20 Recognizes Mitochondrial Presequences Through Dynamic Equilibrium Among Multiple Bound States.
Embo J., 26, 2007
|
|
2V1S
 
 | | CRYSTAL STRUCTURE OF RAT TOM20-ALDH PRESEQUENCE COMPLEX | | Descriptor: | ALDEHYDE DEHYDROGENASE, MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20 HOMOLOG | | Authors: | Obita, T, Igura, M, Ose, T, Endo, T, Maenaka, K, Kohda, D. | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Tom20 Recognizes Mitochondrial Presequences Through Dynamic Equilibrium Among Multiple Bound States.
Embo J., 26, 2007
|
|
7C20
 
 | |
7C21
 
 | | Crystal structure of Duvenhage virus phosphoprotein C-terminal domain | | Descriptor: | Phosphoprotein | | Authors: | Sugiyama, A, Jiang, X, Maenaka, K, Yao, M, Ose, T. | | Deposit date: | 2020-05-06 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural comparison of the C-terminal domain of functionally divergent lyssavirus P proteins.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
5XOF
 
 | | Crystal structure of human paired immunoglobulin-like type 2 receptor alpha with synthesized glycopeptide I | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-6)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Paired immunoglobulin-like type 2 receptor alpha, Peptide from Nitric oxide synthase, ... | | Authors: | Furukawa, A, Kakita, K, Yamada, T, Ishizuka, M, Sakamoto, J, Hatori, N, Maeda, N, Ohsaka, F, Saitoh, T, Nomura, T, Kuroki, K, Nambu, H, Arase, H, Matsunaga, H, Anada, M, Ose, T, Hashimoto, S, Maenaka, K. | | Deposit date: | 2017-05-28 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.963 Å) | | Cite: | Structural and thermodynamic analyses reveal critical features of glycopeptide recognition by the human PILR alpha immune cell receptor.
J. Biol. Chem., 292, 2017
|
|
5XO2
 
 | | Crystal structure of human paired immunoglobulin-like type 2 receptor alpha with synthesized glycopeptide II | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-6)-2-acetamido-2,4-dideoxy-alpha-D-xylo-hexopyranose, Paired immunoglobulin-like type 2 receptor alpha, Peptide from Envelope glycoprotein B | | Authors: | Furukawa, A, Kakita, K, Yamada, T, Ishizuka, M, Sakamoto, J, Hatori, N, Maeda, N, Ohsaka, F, Saitoh, T, Nomura, T, Kuroki, K, Nambu, H, Arase, H, Matsunaga, S, Anada, M, Ose, T, Hashimoto, S, Maenaka, K. | | Deposit date: | 2017-05-25 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and thermodynamic analyses reveal critical features of glycopeptide recognition by the human PILR alpha immune cell receptor.
J. Biol. Chem., 292, 2017
|
|
1WOC
 
 | | Crystal structure of PriB | | Descriptor: | Primosomal replication protein n | | Authors: | Shioi, S, Ose, T, Maenaka, K, Abe, Y, Kohda, D, Katayama, T, Ueda, T. | | Deposit date: | 2004-08-13 | | Release date: | 2005-01-25 | | Last modified: | 2012-12-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a biologically functional form of PriB from Escherichia coli reveals a potential single-stranded DNA-binding site
Biochem.Biophys.Res.Commun., 326, 2005
|
|
2D7H
 
 | | Crystal structure of the ccc complex of the N-terminal domain of PriA | | Descriptor: | DNA (5'-D(P*CP*CP*C)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2D7G
 
 | | Crystal structure of the aa complex of the N-terminal domain of PriA | | Descriptor: | DNA (5'-D(P*AP*A)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
