2DCN
 
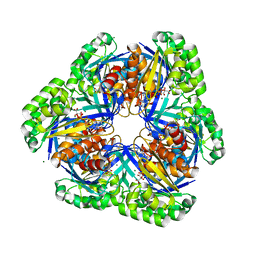 | | Crystal structure of 2-keto-3-deoxygluconate kinase from Sulfolobus tokodaii complexed with 2-keto-6-phosphogluconate (alpha-furanose form) | | Descriptor: | 6-O-phosphono-beta-D-psicofuranosonic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Okazaki, S, Onda, H, Suzuki, A, Kuramitsu, S, Masui, R, Yamane, T. | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of 2-keto-3-deoxygluconate kinase from Sulfolobus tokodaii complexed with 2-keto-6-phosphogluconate
To be Published
|
|
1GEA
 
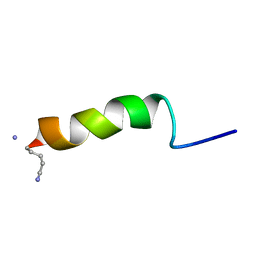 | | RECEPTOR-BOUND CONFORMATION OF PACAP21 | | Descriptor: | PITUITARY ADENYLATE CYCLASE ACTIVATING POLYPEPTIDE | | Authors: | Inooka, H, Ohtaki, T, Kitahara, O, Ikegami, T, Endo, S, Kitada, C, Ogi, K, Onda, H, Fujino, M, Shirakawa, M. | | Deposit date: | 2000-10-20 | | Release date: | 2001-04-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Conformation of a peptide ligand bound to its G-protein coupled receptor.
Nat.Struct.Biol., 8, 2001
|
|
7BW5
 
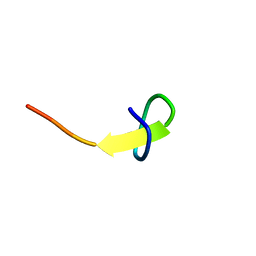 | | a new lasso peptide koreensin | | Descriptor: | lasso peptide koreensin | | Authors: | Hemmi, H, Kodani, S. | | Deposit date: | 2020-04-13 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Heterologous production of new lasso peptide koreensin based on genome mining.
J Antibiot (Tokyo), 74, 2021
|
|
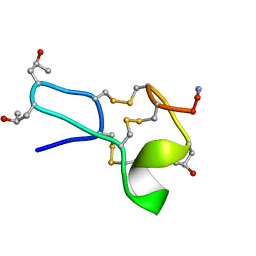
1TCJ
 
 | |
1TCG
 
 | |
1TCK
 
 | |
1TCH
 
 | |
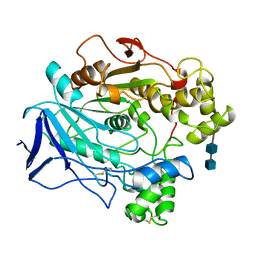
1CRL
 
 | |
1GZ7
 
 | | Crystal structure of the closed state of lipase 2 from Candida rugosa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LIPASE 2 | | Authors: | Mancheno, J.M, Hermoso, J.A. | | Deposit date: | 2002-05-17 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Insights Into the Lipase/Esterase Behavior in the Candida Rugosa Lipases Family: Crystal Structure of the Lipase 2 Isoenzyme at 1.97A Resolution
J.Mol.Biol., 332, 2003
|
|
1TRH
 
 | |
