1PGS
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF PNGASE F, A GLYCOSYLASPARAGINASE FROM FLAVOBACTERIUM MENINGOSEPTICUM | | Descriptor: | PEPTIDE-N(4)-(N-ACETYL-BETA-D-GLUCOSAMINYL)ASPARAGINE AMIDASE F | | Authors: | Norris, G.E, Stillman, T.J, Anderson, B.F, Baker, E.N. | | Deposit date: | 1994-10-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of PNGase F, a glycosylasparaginase from Flavobacterium meningosepticum.
Structure, 2, 1994
|
|
4G89
 
 | | Crystal structure of k. pneumoniae mta/adohcy nucleosidase in complex with fragmented s-adenosyl-l-homocysteine | | Descriptor: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Norris, G.E, Brown, R.L, Anderson, B.F, Tyler, P.C, Evans, G.B. | | Deposit date: | 2012-07-23 | | Release date: | 2012-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of k. pneumoniae mta/adohcy nucleosidase in complex with fragmented s-adenosyl-l-homocysteine
To be Published
|
|
1LFH
 
 | | MOLECULAR REPLACEMENT SOLUTION OF THE STRUCTURE OF APOLACTOFERRIN, A PROTEIN DISPLAYING LARGE-SCALE CONFORMATIONAL CHANGE | | Descriptor: | CHLORIDE ION, LACTOFERRIN | | Authors: | Anderson, B.F, Baker, E.N, Norris, G.E. | | Deposit date: | 1991-09-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular replacement solution of the structure of apolactoferrin, a protein displaying large-scale conformational change.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
2AZA
 
 | |
4YI7
 
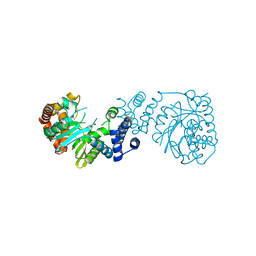 | |
6U1O
 
 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2019-08-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 595, 2021
|
|
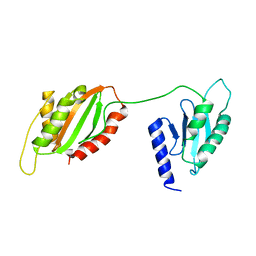
6U1L
 
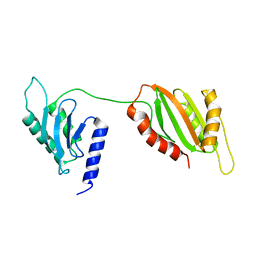 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2019-08-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 595, 2021
|
|
6BQI
 
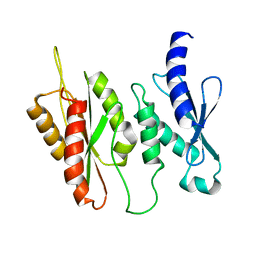 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 2020
|
|
4K7C
 
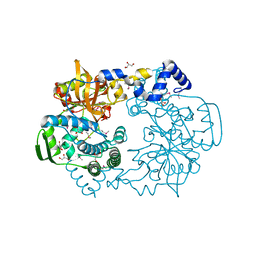 | |
4IUW
 
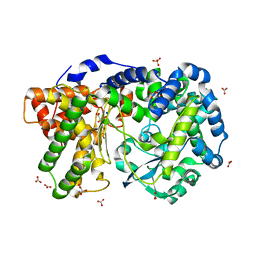 | | Crystal structure of PEPO from Lactobacillus rhamnosis HN001 (DR20) | | Descriptor: | CARBONATE ION, CITRIC ACID, Neutral endopeptidase, ... | | Authors: | Anderson, B.F, Knapp, K.M, Holland, R, Norris, G.E, Christensson, C, Bratt, H, Collins, L.J, Coolbear, T, Lubbers, M.W, Toole, P.W.O, Reid, J.R, Jameson, G.B. | | Deposit date: | 2013-01-21 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of PEPO from Lactobacillus rhamnosis HN001 (DR20)
TO BE PUBLISHED
|
|
4ROT
 
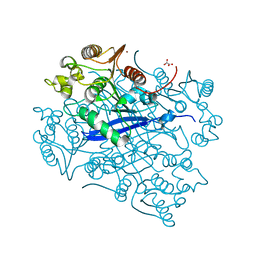 | | Crystal structure of esterase A from Streptococcus pyogenes | | Descriptor: | Esterase A, OXALATE ION, ZINC ION | | Authors: | Bennett, M.D, Holland, R, Coulibaly, F, Loo, T.S, Norris, G.E, Anderson, B.F. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of esterase A from Streptococcus pyogenes
To be Published
|
|
4PO3
 
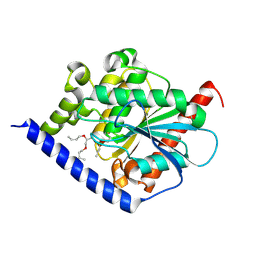 | |
4N5H
 
 | | Crystal structure of ESTERASE B from Lactobacillus Rhamnosis (HN001) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Esterase/lipase, ... | | Authors: | Bennett, M.D, Holland, R, Loo, T.S, Smith, C.A, Norris, G.E, Delabre, M.-L, Anderson, B.F. | | Deposit date: | 2013-10-09 | | Release date: | 2014-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of ESTERASE B from Lactobacillus Rhamnosis (HN001)
TO BE PUBLISHED
|
|
3PMS
 
 | |
4OUK
 
 | | Crystal structure of a C6-C4 SN3 inhibited ESTERASE B from LACTOBACILLUS RHAMNOSIS | | Descriptor: | (2R)-2,3-bis(hexyloxy)propyl hydrogen (S)-pentylphosphonate, Esterase B | | Authors: | Colbert, D.A, Bennett, M.D, Lun, D.J, Holland, R, Delabre, M.-L, Loo, T.S, Anderson, B.F, Norris, G.E. | | Deposit date: | 2014-02-17 | | Release date: | 2014-08-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a C6-C4 SN3 inhibited ESTERASE B from LACTOBACILLUS RHAMNOSIS
TO BE PUBLISHED
|
|
2MVI
 
 | | Structure of the S-glycosylated bacteriocin ASM1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bacteriocin plantarican ASM1 | | Authors: | Goroncy, A.K, Loo, T.S, Koolard, A, Patchett, M.L, Norris, G.E. | | Deposit date: | 2014-10-06 | | Release date: | 2014-12-10 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the S-glycosylated bacteriocin ASM1 from Lactobacillus plantarum
To be Published
|
|
4G41
 
 | | Crystal structure of s-adenosylhomocysteine nucleosidase from streptococcus pyogenes in complex with 5-methylthiotubericidin | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MTA/SAH nucleosidase | | Authors: | Ponniah, K, Norris, G.E, Anderson, B.F, Brown, R.L, Tyler, P.C, Evans, G.B, Frohlich, R. | | Deposit date: | 2012-07-15 | | Release date: | 2012-09-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of s-adenosylhomocysteine nucleosidase from streptococcus pyogenes in complex with 5-methylthiotubericidin;
TO BE PUBLISHED
|
|
4FFS
 
 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Helicobacter pylori with butyl-thio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, CHLORIDE ION, MTA/SAH nucleosidase | | Authors: | Haapalainen, A.M, Rinaldo-Matthis, A, Brown, R.L, Norris, G.E, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-06-01 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Picomolar Transition State Analogue Inhibitor of MTAN as a Specific Antibiotic for Helicobacter pylori.
Biochemistry, 51, 2012
|
|
4GMH
 
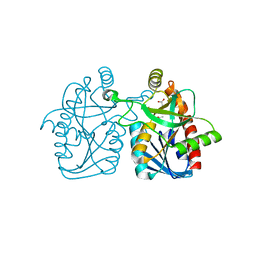 | | Crystal structure of staphylococcus aureus 5'-methylthioadenosine/s-adenosylhomocysteine nucleosidase | | Descriptor: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ACETATE ION | | Authors: | Brown, R.L, Anderson, B.F, Norris, G.E, Tyler, P.C, Evans, G.B, Sutherland-Smith, A.J. | | Deposit date: | 2012-08-15 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of staphylococcus aureus 5'-methylthioadenosine/s-adenosylhomocysteine nucleosidase
To be Published
|
|
4GTN
 
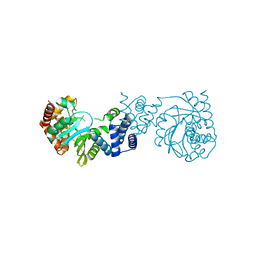 | |
2AKQ
 
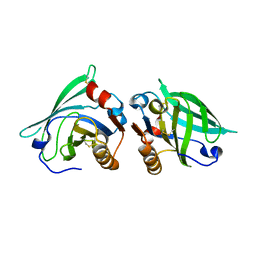 | | The structure of bovine B-lactoglobulin A in crystals grown at very low ionic strength | | Descriptor: | Beta-lactoglobulin variant A | | Authors: | Adams, J.J, Anderson, B.F, Norris, G.E, Creamer, L.K, Jameson, G.B. | | Deposit date: | 2005-08-03 | | Release date: | 2005-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of bovine beta-lactoglobulin (variant A) at very low ionic strength
J.Struct.Biol., 154, 2006
|
|
1ZCO
 
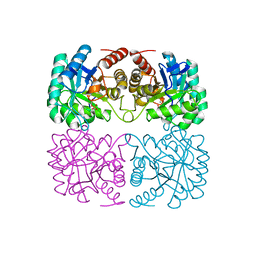 | | Crystal structure of pyrococcus furiosus 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase | | Descriptor: | 2-dehydro-3-deoxyphosphoheptonate aldolase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Schofield, L.R, Anderson, B.F, Patchett, M.L, Norris, G.E, Jameson, G.B, Parker, E.J. | | Deposit date: | 2005-04-12 | | Release date: | 2005-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate Ambiguity and Crystal Structure of Pyrococcus furiosus 3-Deoxy-d-arabino-heptulosonate-7-phosphate Synthase: An Ancestral 3-Deoxyald-2-ulosonate-phosphate Synthase?(,)
Biochemistry, 44, 2005
|
|
3DYV
 
 | |
3E1G
 
 | |
3DLT
 
 | |
