8JBD
 
 | |
6JO3
 
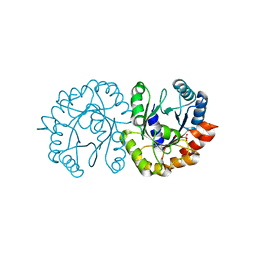 | | Crystal structure of (S)-3-O-geranylgeranylglyceryl phosphate synthase from Thermoplasma acidophilum in complex with substrate sn-glycerol-1-phosphate | | Descriptor: | Geranylgeranylglyceryl phosphate synthase, SN-GLYCEROL-1-PHOSPHATE | | Authors: | Nemoto, N, Miyazono, K, Tanokura, M, Yamagishi, A. | | Deposit date: | 2019-03-20 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of (S)-3-O-geranylgeranylglyceryl phosphate synthase from Thermoplasma acidophilum in complex with the substrate sn-glycerol 1-phosphate.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
3VVU
 
 | | Crystal structure of reconstructed bacterial ancestral NDK, Bac1 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Nemoto, N, Miyazono, K, Kimura, M, Yokobori, S, Akanuma, S, Tanokura, M, Yamagishi, A. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Experimental evidence for the thermophilicity of ancestral life
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3VVT
 
 | | Crystal structure of reconstructed archaeal ancestral NDK, Arc1 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Nemoto, N, Miyazono, K, Kimura, M, Yokobori, S, Akanuma, S, Tanokura, M, Yamagishi, A. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Experimental evidence for the thermophilicity of ancestral life
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6LN3
 
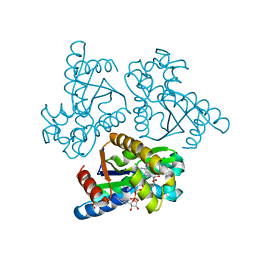 | | Crystal structure of adenylate kinase from an extremophilic archaeon Aeropyrum pernix with ATP and AMP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shibanuma, Y, Nemoto, N, Yamamoto, N, Sampei, G, Kawai, G. | | Deposit date: | 2019-12-28 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of adenylate kinase from an extremophilic archaeon Aeropyrum pernix with ATP and AMP.
J.Biochem., 168, 2020
|
|
1WIC
 
 | | Solution structure of the MSP domain of RIKEN cDNA 6030424E15 | | Descriptor: | Hypothetical Protein RIKEN cDNA 6030424E15 | | Authors: | Endo, H, Asakura, K, Nemoto, N, Takasugi, K, Izumie, K, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the MSP domain of RIKEN cDNA 6030424E15
TO BE PUBLISHED
|
|
1WFH
 
 | | Solution structrue of the zf-AN1 domain from Arabidopsis thaliana At2g36320 protein | | Descriptor: | ZINC ION, zinc finger (AN1-like) family protein | | Authors: | Tomizawa, T, Kigawa, T, Nemoto, N, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-26 | | Release date: | 2004-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structrue of the zf-AN1 domain from Arabidopsis thaliana At2g36320 protein
To be Published
|
|
1WHD
 
 | | Solution structure of the PDZ domain of RGS3 | | Descriptor: | regulator of G-protein signaling 3 | | Authors: | Nakanishi, T, Nemoto, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PDZ domain of RGS3
To be Published
|
|
1IX5
 
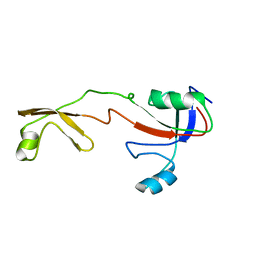 | | Solution structure of the Methanococcus thermolithotrophicus FKBP | | Descriptor: | FKBP | | Authors: | Suzuki, R, Nagata, K, Kawakami, M, Nemoto, N, Furutani, M, Adachi, K, Maruyama, T, Tanokura, M. | | Deposit date: | 2002-06-12 | | Release date: | 2003-06-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional Solution Structure of an Archaeal FKBP with a Dual Function of Peptidyl Prolyl cis-trans Isomerase and Chaperone-like Activities
J.MOL.BIOL., 328, 2003
|
|
1IY3
 
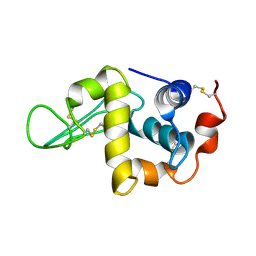 | | Solution Structure of the Human lysozyme at 4 degree C | | Descriptor: | Lysozyme | | Authors: | Kumeta, H, Miura, A, Kobashigawa, Y, Miura, K, Oka, C, Nitta, K, Nemoto, N, Tsuda, S. | | Deposit date: | 2002-07-15 | | Release date: | 2002-07-31 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in human lysozyme elucidated by three-dimensional NMR spectroscopy
Biochemistry, 42, 2003
|
|
1IY4
 
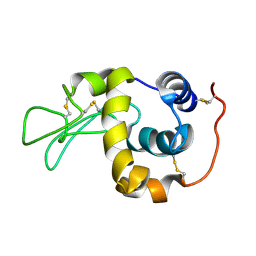 | | Solution structure of the human lysozyme at 35 degree C | | Descriptor: | Lysozyme | | Authors: | Kumeta, H, Miura, A, Kobashigawa, Y, Miura, K, Oka, C, Nitta, K, Nemoto, N, Tsuda, S. | | Deposit date: | 2002-07-15 | | Release date: | 2002-07-31 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in human lysozyme elucidated by three-dimensional NMR spectroscopy
Biochemistry, 42, 2003
|
|
1WFP
 
 | | Solution structure of the zf-AN1 domain from Arabiopsis thaliana F5O11.17 protein | | Descriptor: | ZINC ION, zinc finger (AN1-like) family protein | | Authors: | Tomizawa, T, Kigawa, T, Nemoto, N, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-26 | | Release date: | 2004-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the zf-AN1 domain from Arabiopsis thaliana F5O11.17 protein
To be Published
|
|
1IW4
 
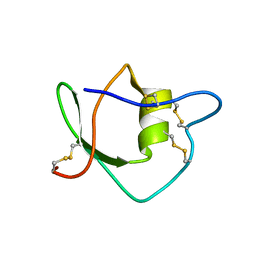 | | Solution structure of ascidian trypsin inhibitor | | Descriptor: | trypsin inhibitor | | Authors: | Hemmi, H, Yoshida, T, Kumazaki, T, Nemoto, N, Hasegawa, J, Nishioka, F, Kyogoku, Y, Yokosawa, H, Kobayashi, Y. | | Deposit date: | 2002-04-19 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ascidian trypsin inhibitor determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 41, 2002
|
|
1WST
 
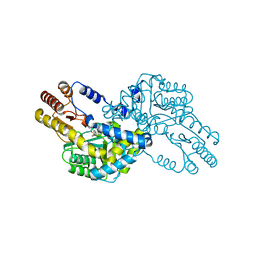 | | Crystal structure of multiple substrate aminotransferase (MsAT) from Thermococcus profundus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, multiple substrate aminotransferase | | Authors: | Lee, W.C, Manabe, F, Nemoto, N, Tamakoshi, M, Tanokura, M, Yamagishi, A. | | Deposit date: | 2004-11-10 | | Release date: | 2005-10-25 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of multiple substrate aminotransferase (MsAT) from Thermococcus profundus
To be Published
|
|
1C8A
 
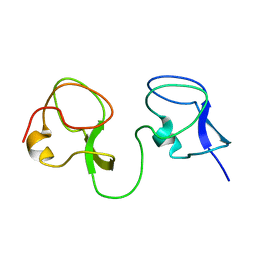 | |
1C89
 
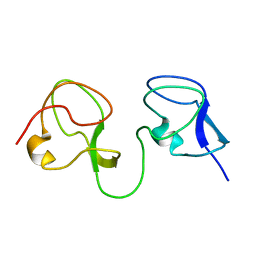 | |
7FG2
 
 | |
7FG3
 
 | |
7FG7
 
 | |
3NLA
 
 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, 40 STRUCTURES | | Descriptor: | ANTIFREEZE PROTEIN RD3 TYPE III | | Authors: | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
3RDN
 
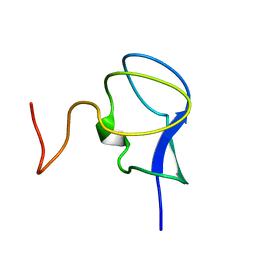 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | ANTIFREEZE PROTEIN RD3 TYPE III | | Authors: | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
