2OIT
 
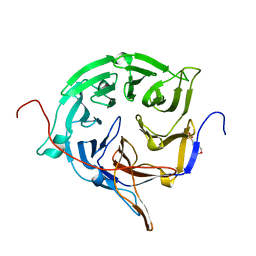 | |
3FMP
 
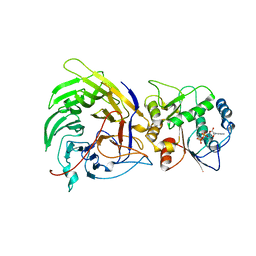 | | Crystal structure of the nucleoporin Nup214 in complex with the DEAD-box helicase Ddx19 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DDX19B, Nuclear pore complex protein Nup214 | | Authors: | Napetschnig, J, Debler, E.W, Blobel, G, Hoelz, A. | | Deposit date: | 2008-12-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FMO
 
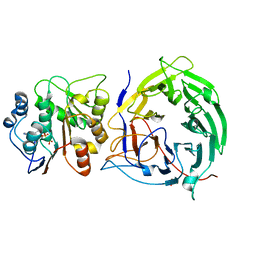 | | Crystal structure of the nucleoporin Nup214 in complex with the DEAD-box helicase Ddx19 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DDX19B, GLYCEROL, ... | | Authors: | Napetschnig, J, Debler, E.W, Blobel, G, Hoelz, A. | | Deposit date: | 2008-12-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1RJ9
 
 | | Structure of the heterodimer of the conserved GTPase domains of the Signal Recognition Particle (Ffh) and Its Receptor (FtsY) | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Signal Recognition Protein, ... | | Authors: | Egea, P.F, Shan, S.O, Napetschnig, J, Savage, D.F, Walter, P, Stroud, R.M. | | Deposit date: | 2003-11-18 | | Release date: | 2004-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate twinning activates the signal recognition particle and its receptor
Nature, 427, 2004
|
|
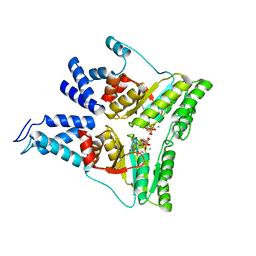
3DLU
 
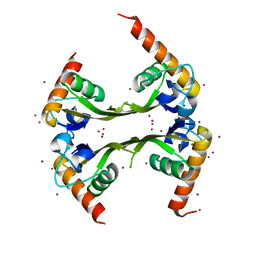 | | Structures of SRP54 and SRP19, the two proteins assembling the ribonucleic core of the Signal Recognition Particle from the archaeon Pyrococcus furiosus. | | Descriptor: | BROMIDE ION, MALONATE ION, Signal recognition particle 19 kDa protein | | Authors: | Egea, P.F, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-29 | | Release date: | 2008-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of SRP54 and SRP19, the two proteins that organize the ribonucleic core of the signal recognition particle from Pyrococcus furiosus.
Plos One, 3, 2008
|
|
3DM9
 
 | | Structures and Conformations in Solution of the Signal Recognition Particle Receptor from the archaeon Pyrococcus furiosus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, Signal recognition particle receptor | | Authors: | Egea, P.F, Tsuruta, H, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-30 | | Release date: | 2008-11-11 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the Signal Recognition Particle Receptor from the Archaeon Pyrococcus furiosus: Implications for the Targeting Step at the Membrane.
Plos One, 3, 2008
|
|
3DLV
 
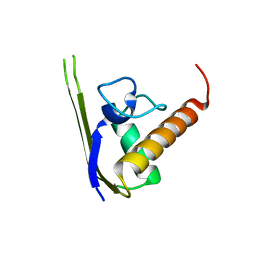 | | Structures of SRP54 and SRP19, the two proteins assembling the ribonucleic core of the Signal Recognition Particle from the archaeon Pyrococcus furiosus. | | Descriptor: | Signal recognition particle 19 kDa protein | | Authors: | Egea, P.F, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-29 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structures of SRP54 and SRP19, the two proteins that organize the ribonucleic core of the signal recognition particle from Pyrococcus furiosus.
Plos One, 3, 2008
|
|
3DM5
 
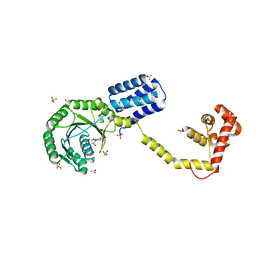 | | Structures of SRP54 and SRP19, the two proteins assembling the ribonucleic core of the Signal Recognition Particle from the archaeon Pyrococcus furiosus. | | Descriptor: | ACETATE ION, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Egea, P.F, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-30 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structures of SRP54 and SRP19, the two proteins that organize the ribonucleic core of the signal recognition particle from Pyrococcus furiosus.
Plos One, 3, 2008
|
|
3DMD
 
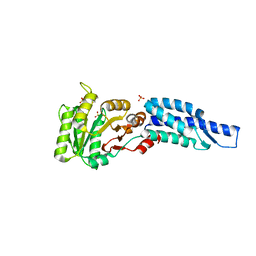 | | Structures and Conformations in Solution of the Signal Recognition Particle Receptor from the archaeon Pyrococcus furiosus | | Descriptor: | GLYCEROL, SULFATE ION, Signal recognition particle receptor | | Authors: | Egea, P.F, Tsuruta, H, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-30 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of the Signal Recognition Particle Receptor from the Archaeon Pyrococcus furiosus: Implications for the Targeting Step at the Membrane.
Plos One, 3, 2008
|
|
3E70
 
 | | Structures and conformations in solution of the Signal Recognition Particle Receptor from the Archaeon Pyrococcus Furiosus | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Signal recognition particle receptor | | Authors: | Egea, P.F, Tsuruta, H, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-08-17 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of the signal recognition particle receptor from the archaeon Pyrococcus furiosus: implications for the targeting step at the membrane.
Plos One, 3, 2008
|
|
4GA0
 
 | |
4GA2
 
 | |
4GA1
 
 | |
